Estimating T-cell repertoire diversity: limitations of classical estimators and a new approach
- PMID: 26150657
- PMCID: PMC4528489
- DOI: 10.1098/rstb.2014.0291
Estimating T-cell repertoire diversity: limitations of classical estimators and a new approach
Abstract
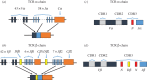
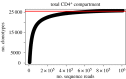
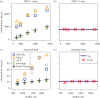
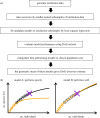
A highly diverse T-cell receptor (TCR) repertoire is a fundamental property of an effective immune system, and is associated with efficient control of viral infections and other pathogens. However, direct measurement of total TCR diversity is impossible. The diversity is high and the frequency distribution of individual TCRs is heavily skewed; the diversity therefore cannot be captured in a blood sample. Consequently, estimators of the total number of TCR clonotypes that are present in the individual, in addition to those observed, are essential. This is analogous to the 'unseen species problem' in ecology. We review the diversity (species richness) estimators that have been applied to T-cell repertoires and the methods used to validate these estimators. We show that existing approaches have significant shortcomings, and frequently underestimate true TCR diversity. We highlight our recently developed estimator, DivE, which can accurately estimate diversity across a range of immunological and biological systems.
Keywords: T-cell receptor repertoire; diversity; species richness.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

