The role of the local microbial ecosystem in respiratory health and disease
- PMID: 26150660
- PMCID: PMC4528492
- DOI: 10.1098/rstb.2014.0294
The role of the local microbial ecosystem in respiratory health and disease
Abstract
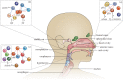
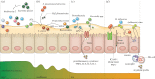
Respiratory tract infections are a major global health concern, accounting for high morbidity and mortality, especially in young children and elderly individuals. Traditionally, highly common bacterial respiratory tract infections, including otitis media and pneumonia, were thought to be caused by a limited number of pathogens including Streptococcus pneumoniae and Haemophilus influenzae. However, these pathogens are also frequently observed commensal residents of the upper respiratory tract (URT) and form-together with harmless commensal bacteria, viruses and fungi-intricate ecological networks, collectively known as the 'microbiome'. Analogous to the gut microbiome, the respiratory microbiome at equilibrium is thought to be beneficial to the host by priming the immune system and providing colonization resistance, while an imbalanced ecosystem might predispose to bacterial overgrowth and development of respiratory infections. We postulate that specific ecological perturbations of the bacterial communities in the URT can occur in response to various lifestyle or environmental effectors, leading to diminished colonization resistance, loss of containment of newly acquired or resident pathogens, preluding bacterial overgrowth, ultimately resulting in local or systemic bacterial infections. Here, we review the current body of literature regarding niche-specific upper respiratory microbiota profiles within human hosts and the changes occurring within these profiles that are associated with respiratory infections.
Keywords: bacterial interaction; colonization resistance; microbiome; pathogenesis; respiratory infections; upper respiratory tract.
© 2015 The Author(s) Published by the Royal Society. All rights reserved.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
