Using linkage maps to correct and scaffold de novo genome assemblies: methods, challenges, and computational tools
- PMID: 26150829
- PMCID: PMC4473057
- DOI: 10.3389/fgene.2015.00220
Using linkage maps to correct and scaffold de novo genome assemblies: methods, challenges, and computational tools
Abstract
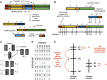
Modern high-throughput DNA sequencing has made it possible to inexpensively produce genome sequences, but in practice many of these draft genomes are fragmented and incomplete. Genetic linkage maps based on recombination rates between physical markers have been used in biology for over 100 years and a linkage map, when paired with a de novo sequencing project, can resolve mis-assemblies and anchor chromosome-scale sequences. Here, I summarize the methodology behind integrating de novo assemblies and genetic linkage maps, outline the current challenges, review the available software tools, and discuss new mapping technologies.
Keywords: draft genome; next-generation sequencing; optical mapping; physical mapping; scaffolds.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

