PubChem structure-activity relationship (SAR) clusters
- PMID: 26150895
- PMCID: PMC4492103
- DOI: 10.1186/s13321-015-0070-x
PubChem structure-activity relationship (SAR) clusters
Abstract
Background: Developing structure-activity relationships (SARs) of molecules is an important approach in facilitating hit exploration in the early stage of drug discovery. Although information on millions of compounds and their bioactivities is freely available to the public, it is very challenging to infer a meaningful and novel SAR from that information.
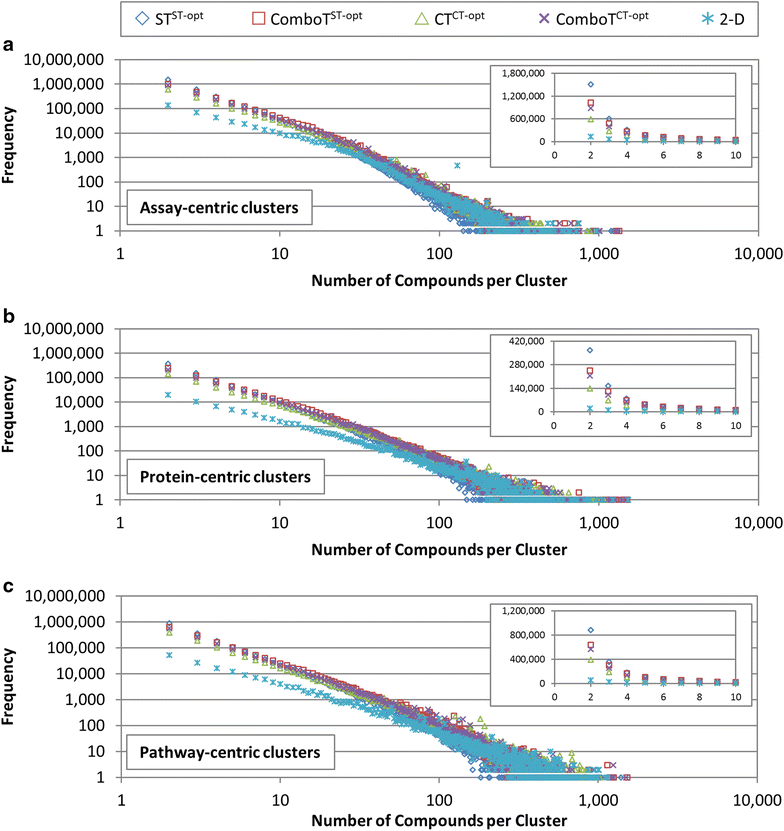
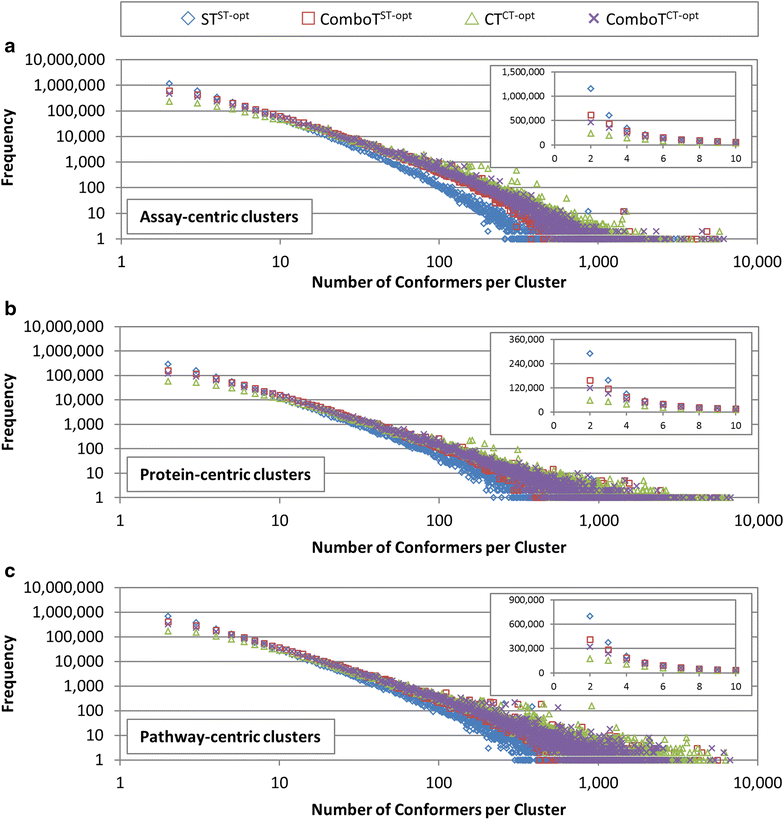
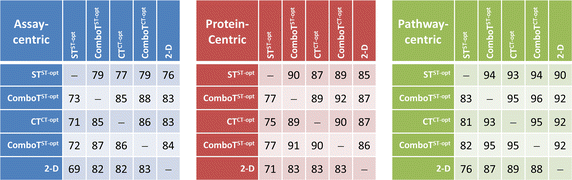
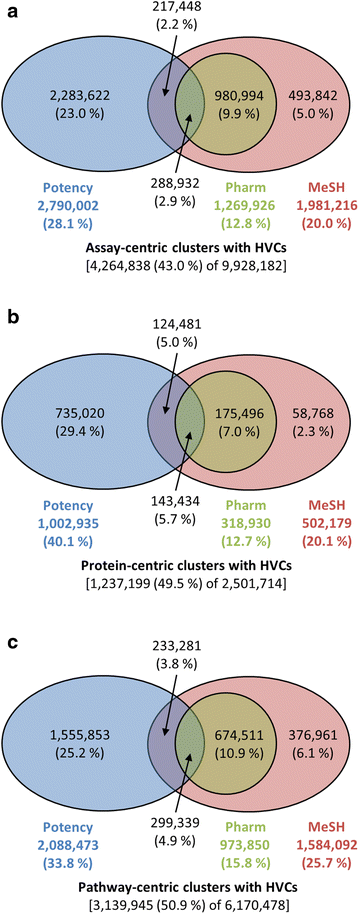
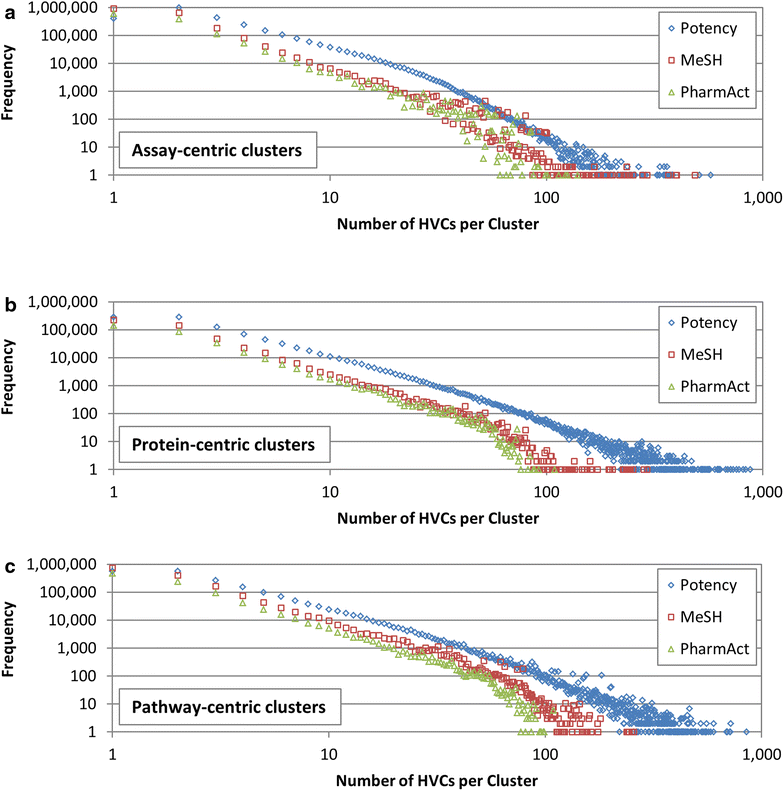
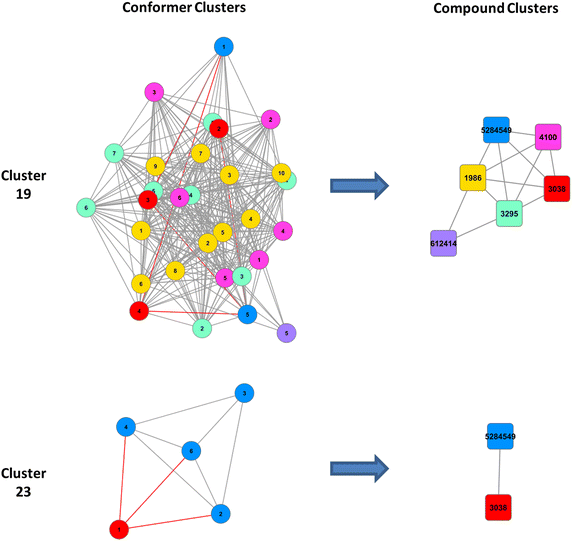
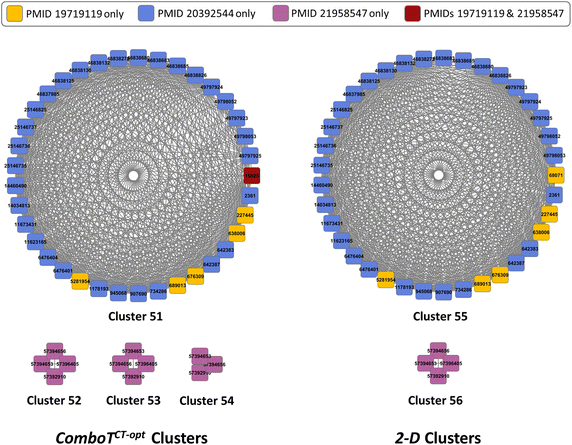
Results: Research discussed in the present paper employed a bioactivity-centered clustering approach to group 843,845 non-inactive compounds stored in PubChem according to both structural similarity and bioactivity similarity, with the aim of mining bioactivity data in PubChem for useful SAR information. The compounds were clustered in three bioactivity similarity contexts: (1) non-inactive in a given bioassay, (2) non-inactive against a given protein, and (3) non-inactive against proteins involved in a given pathway. In each context, these small molecules were clustered according to their two-dimensional (2-D) and three-dimensional (3-D) structural similarities. The resulting 18 million clusters, named "PubChem SAR clusters", were delivered in such a way that each cluster contains a group of small molecules similar to each other in both structure and bioactivity.
Conclusions: The PubChem SAR clusters, pre-computed using publicly available bioactivity information, make it possible to quickly navigate and narrow down the compounds of interest. Each SAR cluster can be a useful resource in developing a meaningful SAR or enable one to design or expand compound libraries from the cluster. It can also help to predict the potential therapeutic effects and pharmacological actions of less-known compounds from those of well-known compounds (i.e., drugs) in the same cluster.
Keywords: BioSystems; Cluster analysis; MeSH; Molecular similarity; PubChem; PubChem3D; Structure–activity relationship (SAR).
Figures




References
-
- Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. In: Ralph AW, David CS, editors. Annual reports in computational chemistry. Amsterdam: Elsevier; 2008. pp. 217–241.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

