Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III
- PMID: 26151409
- PMCID: PMC4506509
- DOI: 10.1038/ncomms8623
Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III
Abstract
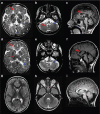
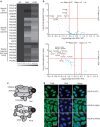
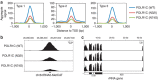
A small proportion of 4H (Hypomyelination, Hypodontia and Hypogonadotropic Hypogonadism) or RNA polymerase III (POLR3)-related leukodystrophy cases are negative for mutations in the previously identified causative genes POLR3A and POLR3B. Here we report eight of these cases carrying recessive mutations in POLR1C, a gene encoding a shared POLR1 and POLR3 subunit, also mutated in some Treacher Collins syndrome (TCS) cases. Using shotgun proteomics and ChIP sequencing, we demonstrate that leukodystrophy-causative mutations, but not TCS mutations, in POLR1C impair assembly and nuclear import of POLR3, but not POLR1, leading to decreased binding to POLR3 target genes. This study is the first to show that distinct mutations in a gene coding for a shared subunit of two RNA polymerases lead to selective modification of the enzymes' availability leading to two different clinical conditions and to shed some light on the pathophysiological mechanism of one of the most common hypomyelinating leukodystrophies, POLR3-related leukodystrophy.
Figures


References
-
- Schiffmann R. & van der Knaap M. S. The latest on leukodystrophies. Curr. Opin. Neurol. 17, 187–192 (2004). - PubMed
-
- Bernard G. & Vanderver A. in GeneReviews® (eds Pagon, R. A., Adam, M. P., Ardinger, H. H., et al..) (University of Washington, Seattle, 1993–2015), http://www.ncbi.nlm.nih.gov/books/NBK99167/. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

