Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae
- PMID: 26151772
- PMCID: PMC4615593
- DOI: 10.1080/15476286.2015.1058477
Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae
Abstract
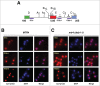
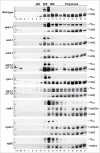
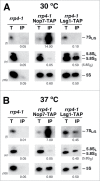
Evolution has provided eukaryotes with mechanisms that impede immature and/or aberrant ribosomes to engage in translation. These mechanisms basically either prevent the nucleo-cytoplasmic export of these particles or, once in the cytoplasm, the release of associated assembly factors, which interfere with the binding of translation initiation factors and/or the ribosomal subunit joining. We have previously shown that aberrant yeast 40S ribosomal subunits containing the 20S pre-rRNA can engage in translation. In this study, we describe that cells harbouring the dob1-1 allele, encoding a mutated version of the exosome-assisting RNA helicase Mtr4, accumulate otherwise nuclear pre-60S ribosomal particles containing the 7S pre-rRNA in the cytoplasm. Polysome fractionation analyses revealed that these particles are competent for translation and do not induce elongation stalls. This phenomenon is rather specific since most mutations in other exosome components or co-factors, impairing the 3' end processing of the mature 5.8S rRNA, accumulate 7S pre-rRNAs in the nucleus. In addition, we confirm that pre-60S ribosomal particles containing either 5.8S + 30 or 5.8S + 5 pre-rRNAs also engage in translation elongation. We propose that 7S pre-rRNA processing is not strictly required for pre-60S r-particle export and that, upon arrival in the cytoplasm, there is no specific mechanism to prevent translation by premature pre-60S r-particles containing 3' extended forms of mature 5.8S rRNA.
Keywords: DAPI, 4,6-diamidino-2-phenylindole; FISH, fluorescence in situ hybridization; Mtr4/Dob1; NRD, Non-functional rRNA decay; RNA exosome; RNA helicase; Ribosome biogenesis; TRAMP complexes, Trf/Air/Mtr4 complexes; Translation; pre-rRNA processing; pre-rRNA, precursor rRNA; r-particles, ribosomal particles; r-proteins, ribosomal proteins; r-subunits, ribosomal subunits; yeast.
Figures



Similar articles
-
Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae.EMBO J. 2010 Jan 6;29(1):80-92. doi: 10.1038/emboj.2009.307. Epub 2009 Nov 5. EMBO J. 2010. PMID: 19893492 Free PMC article.
-
The ribosome assembly factor Nop53 controls association of the RNA exosome with pre-60S particles in yeast.J Biol Chem. 2019 Dec 13;294(50):19365-19380. doi: 10.1074/jbc.RA119.010193. Epub 2019 Oct 29. J Biol Chem. 2019. PMID: 31662437 Free PMC article.
-
Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits.Nat Struct Mol Biol. 2012 Aug;19(8):744-53. doi: 10.1038/nsmb.2308. Epub 2012 Jul 1. Nat Struct Mol Biol. 2012. PMID: 22751017 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Maturation of pre-40S particles in yeast and humans.Wiley Interdiscip Rev RNA. 2019 Jan;10(1):e1516. doi: 10.1002/wrna.1516. Epub 2018 Nov 8. Wiley Interdiscip Rev RNA. 2019. PMID: 30406965 Review.
Cited by
-
An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression.Genes Dev. 2017 Jun 15;31(12):1257-1271. doi: 10.1101/gad.302604.117. Epub 2017 Jul 21. Genes Dev. 2017. PMID: 28733371 Free PMC article.
-
Nucleolar localization of the yeast RNA exosome subunit Rrp44 hints at early pre-rRNA processing as its main function.J Biol Chem. 2020 Aug 7;295(32):11195-11213. doi: 10.1074/jbc.RA120.013589. Epub 2020 Jun 17. J Biol Chem. 2020. PMID: 32554806 Free PMC article.
-
A stable XPG protein is required for proper ribosome biogenesis: Insights on the phenotype of combinate Xeroderma Pigmentosum/Cockayne Syndrome patients.PLoS One. 2022 Jul 8;17(7):e0271246. doi: 10.1371/journal.pone.0271246. eCollection 2022. PLoS One. 2022. PMID: 35802638 Free PMC article.
-
Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases.Biomolecules. 2018 Oct 24;8(4):123. doi: 10.3390/biom8040123. Biomolecules. 2018. PMID: 30356013 Free PMC article. Review.
-
Cooling-induced SUMOylation of EXOSC10 down-regulates ribosome biogenesis.RNA. 2016 Apr;22(4):623-35. doi: 10.1261/rna.054411.115. Epub 2016 Feb 8. RNA. 2016. PMID: 26857222 Free PMC article.
References
-
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 2008; 65:2334-59; PMID:18408888; http://dx.doi.org/10.1007/s00018-008-8027-0 - DOI - PMC - PubMed
-
- Woolford JL Jr., Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 2013; 195:643-81; PMID:24190922; http://dx.doi.org/10.1534/genetics.113.153197 - DOI - PMC - PubMed
-
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene 2003; 313:17-42; PMID:12957375; http://dx.doi.org/10.1016/S0378-1119(03)00629-2 - DOI - PubMed
-
- de la Cruz J, Kressler D, Linder P. Ribosomal subunit assembly In: Olson MOJ, ed. Nucleolus. Georgetown: Kluwer academic; LandesBioscience/eurekah.com, 2004:258-85
-
- Gerhardy S, Menet AM, Pena C, Petkowski JJ, Panse VG. Assembly and nuclear export of pre-ribosomal particles in budding yeast. Chromosoma 2014; 123:327-44; PMID:24817020; http://dx.doi.org/10.1007/s00412-014-0463-z - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
