Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription
- PMID: 26151904
- PMCID: PMC4495351
- DOI: 10.1016/j.devcel.2015.05.012
Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription
Abstract
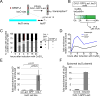
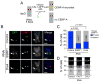
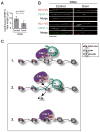
Centromeres are essential chromosomal structures that mediate accurate chromosome segregation during cell division. Centromeres are specified epigenetically by the heritable incorporation of the centromeric histone H3 variant CENP-A. While many of the primary factors that mediate centromeric deposition of CENP-A are known, the chromatin and DNA requirements of this process have remained elusive. Here, we uncover a role for transcription in Drosophila CENP-A deposition. Using an inducible ectopic centromere system that uncouples CENP-A deposition from endogenous centromere function and cell-cycle progression, we demonstrate that CENP-A assembly by its loading factor, CAL1, requires RNAPII-mediated transcription of the underlying DNA. This transcription depends on the CAL1 binding partner FACT, but not on CENP-A incorporation. Our work establishes RNAPII passage as a key step in chaperone-mediated CENP-A chromatin establishment and propagation.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures




Similar articles
-
The checkpoint protein Zw10 connects CAL1-dependent CENP-A centromeric loading and mitosis duration in Drosophila cells.PLoS Genet. 2019 Sep 25;15(9):e1008380. doi: 10.1371/journal.pgen.1008380. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31553715 Free PMC article.
-
Co-evolving CENP-A and CAL1 Domains Mediate Centromeric CENP-A Deposition across Drosophila Species.Dev Cell. 2016 Apr 18;37(2):136-47. doi: 10.1016/j.devcel.2016.03.021. Dev Cell. 2016. PMID: 27093083 Free PMC article.
-
The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C.PLoS Biol. 2012;10(12):e1001460. doi: 10.1371/journal.pbio.1001460. Epub 2012 Dec 27. PLoS Biol. 2012. PMID: 23300382 Free PMC article.
-
A network of players in H3 histone variant deposition and maintenance at centromeres.Biochim Biophys Acta. 2014 Mar;1839(3):241-50. doi: 10.1016/j.bbagrm.2013.11.008. Epub 2013 Dec 6. Biochim Biophys Acta. 2014. PMID: 24316467 Review.
-
Centromere regulation: new players, new rules, new questions.Eur J Cell Biol. 2011 Oct;90(10):805-10. doi: 10.1016/j.ejcb.2011.04.016. Eur J Cell Biol. 2011. PMID: 21684630 Review.
Cited by
-
Centromeric Transcription: A Conserved Swiss-Army Knife.Genes (Basel). 2020 Aug 9;11(8):911. doi: 10.3390/genes11080911. Genes (Basel). 2020. PMID: 32784923 Free PMC article. Review.
-
Centromeres Transcription and Transcripts for Better and for Worse.Prog Mol Subcell Biol. 2021;60:169-201. doi: 10.1007/978-3-030-74889-0_7. Prog Mol Subcell Biol. 2021. PMID: 34386876
-
Centromeric Non-coding Transcription: Opening the Black Box of Chromosomal Instability?Curr Genomics. 2017 Jun;18(3):227-235. doi: 10.2174/1389202917666161102095508. Curr Genomics. 2017. PMID: 28603453 Free PMC article. Review.
-
GST-IVTT pull-down: a fast and versatile in vitro method for validating and mapping protein-protein interactions.FEBS Open Bio. 2022 Nov;12(11):1988-1995. doi: 10.1002/2211-5463.13485. Epub 2022 Sep 22. FEBS Open Bio. 2022. PMID: 36102272 Free PMC article.
-
Two-Color CRISPR Imaging Reveals Dynamics of Herpes Simplex Virus 1 Replication Compartments and Virus-Host Interactions.J Virol. 2022 Dec 21;96(24):e0092022. doi: 10.1128/jvi.00920-22. Epub 2022 Dec 1. J Virol. 2022. PMID: 36453882 Free PMC article.
References
-
- Adolph S, Brusselbach S, Muller R. Inhibition of transcription blocks cell cycle progression of NIH3T3 fibroblasts specifically in G1. J Cell Sci. 1993;105 ( Pt 1):113–122. - PubMed
-
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

