Upregulating endogenous genes by an RNA-programmable artificial transactivator
- PMID: 26152305
- PMCID: PMC4652751
- DOI: 10.1093/nar/gkv682
Upregulating endogenous genes by an RNA-programmable artificial transactivator
Abstract
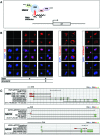
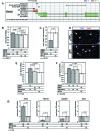
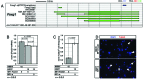
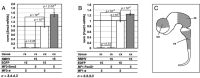
To promote expression of endogenous genes ad libitum, we developed a novel, programmable transcription factor prototype. Kept together via an MS2 coat protein/RNA interface, it includes a fixed, polypeptidic transactivating domain and a variable RNA domain that recognizes the desired gene. Thanks to this device, we specifically upregulated five genes, in cell lines and primary cultures of murine pallial precursors. Gene upregulation was small, however sufficient to robustly inhibit neuronal differentiation. The transactivator interacted with target gene chromatin via its RNA cofactor. Its activity was restricted to cells in which the target gene is normally transcribed. Our device might be useful for specific applications. However for this purpose, it will require an improvement of its transactivation power as well as a better characterization of its target specificity and mechanism of action.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Janowski B.A., Younger S.T., Hardy D.B., Ram R., Huffman K.E., Corey D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. - PubMed
-
- Turunen M.P., Lehtola T., Heinonen S.E., Assefa G.S., Korpisalo P., Girnary R., Glass C.K., Väisänen S., Ylä-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ. Res. 2009;105:604–609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

