NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication
- PMID: 26152585
- PMCID: PMC4488949
- DOI: 10.1128/mBio.00759-15
NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication
Abstract
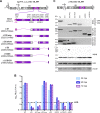
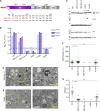
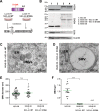
Induction of membrane rearrangements in the cytoplasm of infected cells is a hallmark of positive-strand RNA viruses. These altered membranes serve as scaffolds for the assembly of viral replication factories (RFs). We have recently shown that hepatitis C virus (HCV) infection induces endoplasmic reticulum-derived double-membrane vesicles (DMVs) representing the major constituent of the RF within the infected cell. RF formation requires the concerted action of nonstructural action of nonstructural protein (NS)3, -4A, protein (NS)3 -4A, -4B, -5A, and -5B. Although the sole expression of NS5A is sufficient to induce DMV formation, its efficiency is very low. In this study, we dissected the determinants within NS5A responsible for DMV formation and found that RNA-binding domain 1 (D1) and the amino-terminal membrane anchor are indispensable for this process. In contrast, deletion of NS5A D2 or D3 did not affect DMV formation but disrupted RNA replication and virus assembly, respectively. To identify cis- and trans-acting factors of DMV formation, we established a trans cleavage assay. We found that induction of DMVs requires full-length NS3, whereas a helicase-lacking mutant was unable to trigger DMV formation in spite of efficient polyprotein cleavage. Importantly, a mutation accelerating cleavage kinetics at the NS4B-5A site diminished DMV formation, while the insertion of an internal ribosome entry site mimicking constitutive cleavage at this boundary completely abolished this process. These results identify key determinants governing the biogenesis of the HCV RF with possible implications for our understanding of how RFs are formed in other positive-strand RNA viruses.
Importance: Like all positive-strand RNA viruses, hepatitis C virus (HCV) extensively reorganizes intracellular membranes to allow efficient RNA replication. Double-membrane vesicles (DMVs) that putatively represent sites of HCV RNA amplification are induced by the concerted action of viral and cellular factors. However, the contribution of individual proteins to this process remains poorly understood. Here we identify determinants in the HCV replicase that are required for DMV biogenesis. Major contributors to this process are domain 1 of nonstructural protein 5A and the helicase domain of nonstructural protein 3. In addition, efficient DMV induction depends on cis cleavage of the viral polyprotein, as well as tightly regulated cleavage kinetics. These results identify key determinants governing the biogenesis of the HCV replication factory with possible implications for our understanding of how this central compartment is formed in other positive-strand RNA viruses.
Copyright © 2015 Romero-Brey et al.
Figures





References
-
- Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. doi:10.1371/journal.ppat.1003056. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
