Genomics and Ecophysiology of Heterotrophic Nitrogen-Fixing Bacteria Isolated from Estuarine Surface Water
- PMID: 26152586
- PMCID: PMC4495170
- DOI: 10.1128/mBio.00929-15
Genomics and Ecophysiology of Heterotrophic Nitrogen-Fixing Bacteria Isolated from Estuarine Surface Water
Abstract
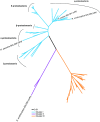
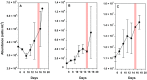
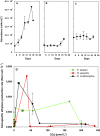
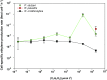
The ability to reduce atmospheric nitrogen (N2) to ammonia, known as N2 fixation, is a widely distributed trait among prokaryotes that accounts for an essential input of new N to a multitude of environments. Nitrogenase reductase gene (nifH) composition suggests that putative N2-fixing heterotrophic organisms are widespread in marine bacterioplankton, but their autecology and ecological significance are unknown. Here, we report genomic and ecophysiology data in relation to N2 fixation by three environmentally relevant heterotrophic bacteria isolated from Baltic Sea surface water: Pseudomonas stutzeri strain BAL361 and Raoultella ornithinolytica strain BAL286, which are gammaproteobacteria, and Rhodopseudomonas palustris strain BAL398, an alphaproteobacterium. Genome sequencing revealed that all were metabolically versatile and that the gene clusters encoding the N2 fixation complex varied in length and complexity between isolates. All three isolates could sustain growth by N2 fixation in the absence of reactive N, and this fixation was stimulated by low concentrations of oxygen in all three organisms (≈ 4 to 40 µmol O2 liter(-1)). P. stutzeri BAL361 did, however, fix N at up to 165 µmol O2 liter(-1), presumably accommodated through aggregate formation. Glucose stimulated N2 fixation in general, and reactive N repressed N2 fixation, except that ammonium (NH4 (+)) stimulated N2 fixation in R. palustris BAL398, indicating the use of nitrogenase as an electron sink. The lack of correlations between nitrogenase reductase gene expression and ethylene (C2H4) production indicated tight posttranscriptional-level control. The N2 fixation rates obtained suggested that, given the right conditions, these heterotrophic diazotrophs could contribute significantly to in situ rates.
Importance: The biological process of importing atmospheric N2 is of paramount importance in terrestrial and aquatic ecosystems. In the oceans, a diverse array of prokaryotes seemingly carry the genetic capacity to perform this process, but lack of knowledge about their autecology and the factors that constrain their N2 fixation hamper an understanding of their ecological importance in marine waters. The present study documents a high variability of genomic and ecophysiological properties related to N2 fixation in three heterotrophic isolates obtained from estuarine surface waters and shows that these organisms fix N2 under a surprisingly broad range of conditions and at significant rates. The observed intricate regulation of N2 fixation for the isolates indicates that indigenous populations of heterotrophic diazotrophs have discrete strategies to cope with environmental controls of N2 fixation. Hence, community-level generalizations about the regulation of N2 fixation in marine heterotrophic bacterioplankton may be problematic.
Copyright © 2015 Bentzon-Tilia et al.
Figures

References
-
- Howarth RW, Marino R, Lane J, Cole JJ. 1988. Nitrogen fixation in freshwater, estuarine, and marine ecosystems. 1. Rates and importance. Limnol Oceanogr 33:669–687. doi:10.4319/lo.1988.33.4_part_2.0669. - DOI
-
- Karl D, Michaels A, Bergman B, Capone D, Carpenter E, Letelier R, Lipschultz F, Paerl H, Sigman D, Stal L. 2002. Dinitrogen fixation in the world’s oceans. Biogeochemistry 57:47–98. doi:10.1023/A:1015798105851. - DOI
-
- Zehr JP, Turner PJ. 2001. Nitrogen fixation: nitrogenase genes and gene expression. Methods Microbiol 30:271–286.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

