The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond
- PMID: 26152727
- PMCID: PMC4543635
- DOI: 10.1074/jbc.R115.656462
The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond
Abstract
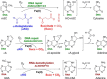
The AlkB family of Fe(II)- and α-ketoglutarate-dependent dioxygenases is a class of ubiquitous direct reversal DNA repair enzymes that remove alkyl adducts from nucleobases by oxidative dealkylation. The prototypical and homonymous family member is an Escherichia coli "adaptive response" protein that protects the bacterial genome against alkylation damage. AlkB has a wide variety of substrates, including monoalkyl and exocyclic bridged adducts. Nine mammalian AlkB homologs exist (ALKBH1-8, FTO), but only a subset functions as DNA/RNA repair enzymes. This minireview presents an overview of the AlkB proteins including recent data on homologs, structural features, substrate specificities, and experimental strategies for studying DNA repair by AlkB family proteins.
Keywords: DNA demethylation; DNA repair; RNA demethylation; RNA repair; alkB; alkylation damage; dioxygenase; direct reversal; metalloprotein; substrate specificity.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

