SWEEP: A Tool for Filtering High-Quality SNPs in Polyploid Crops
- PMID: 26153076
- PMCID: PMC4555216
- DOI: 10.1534/g3.115.019703
SWEEP: A Tool for Filtering High-Quality SNPs in Polyploid Crops
Abstract
High-throughput next-generation sequence-based genotyping and single nucleotide polymorphism (SNP) detection opens the door for emerging genomics-based breeding strategies such as genome-wide association analysis and genomic selection. In polyploids, SNP detection is confounded by a highly similar homeologous sequence where a polymorphism between subgenomes must be differentiated from a SNP. We have developed and implemented a novel tool called SWEEP: Sliding Window Extraction of Explicit Polymorphisms. SWEEP uses subgenome polymorphism haplotypes as contrast to identify true SNPs between genotypes. The tool is a single command script that calls a series of modules based on user-defined options and takes sorted/indexed bam files or vcf files as input. Filtering options are highly flexible and include filtering based on sequence depth, alternate allele ratio, and SNP quality on top of the SWEEP filtering procedure. Using real and simulated data we show that SWEEP outperforms current SNP filtering methods for polyploids. SWEEP can be used for high-quality SNP discovery in polyploid crops.
Keywords: SNP; peanut; polyploidy.
Copyright © 2015 Clevenger and Ozias-Akins.
Figures


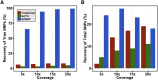
References
-
- Bertioli D. J., Vidigal B., Nielen S., Ratnaparkhe M. B., Lee T. H., et al. , 2013. The repetitive component of the A genome of peanut (Arachis hypogaea) and its role in remodelling intergenic sequence space since its evolutionary divergence from the B genome. Ann. Bot. (Lond.) 112: 545–559. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
