Genome Wide Distributions and Functional Characterization of Copy Number Variations between Chinese and Western Pigs
- PMID: 26154170
- PMCID: PMC4496047
- DOI: 10.1371/journal.pone.0131522
Genome Wide Distributions and Functional Characterization of Copy Number Variations between Chinese and Western Pigs
Abstract
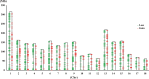
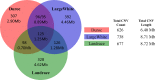
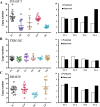
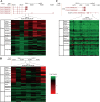
Copy number variations (CNVs) refer to large insertions, deletions and duplications in the genomic structure ranging from one thousand to several million bases in size. Since the development of next generation sequencing technology, several methods have been well built for detection of copy number variations with high credibility and accuracy. Evidence has shown that CNV occurring in gene region could lead to phenotypic changes due to the alteration in gene structure and dosage. However, it still remains unexplored whether CNVs underlie the phenotypic differences between Chinese and Western domestic pigs. Based on the read-depth methods, we investigated copy number variations using 49 individuals derived from both Chinese and Western pig breeds. A total of 3,131 copy number variation regions (CNVRs) were identified with an average size of 13.4 Kb in all individuals during domestication, harboring 1,363 genes. Among them, 129 and 147 CNVRs were Chinese and Western pig specific, respectively. Gene functional enrichments revealed that these CNVRs contribute to strong disease resistance and high prolificacy in Chinese domestic pigs, but strong muscle tissue development in Western domestic pigs. This finding is strongly consistent with the morphologic characteristics of Chinese and Western pigs, indicating that these group-specific CNVRs might have been preserved by artificial selection for the favored phenotypes during independent domestication of Chinese and Western pigs. In this study, we built high-resolution CNV maps in several domestic pig breeds and discovered the group specific CNVs by comparing Chinese and Western pigs, which could provide new insight into genomic variations during pigs' independent domestication, and facilitate further functional studies of CNV-associated genes.
Conflict of interest statement
Figures
Similar articles
-
Improved Detection and Characterization of Copy Number Variations Among Diverse Pig Breeds by Array CGH.G3 (Bethesda). 2015 Apr 22;5(6):1253-61. doi: 10.1534/g3.115.018473. G3 (Bethesda). 2015. PMID: 25908567 Free PMC article.
-
Copy number variation detection using SNP genotyping arrays in three Chinese pig breeds.Anim Genet. 2015 Apr;46(2):101-9. doi: 10.1111/age.12247. Epub 2015 Jan 15. Anim Genet. 2015. PMID: 25590996
-
Identification of genome-wide copy number variations among diverse pig breeds by array CGH.BMC Genomics. 2012 Dec 24;13:725. doi: 10.1186/1471-2164-13-725. BMC Genomics. 2012. PMID: 23265576 Free PMC article.
-
Genome wide copy number variations using Porcine 60K SNP Beadchip in Landlly pigs.Anim Biotechnol. 2023 Nov;34(6):1891-1899. doi: 10.1080/10495398.2022.2056047. Epub 2022 Apr 4. Anim Biotechnol. 2023. PMID: 35369845 Review.
-
Copy number variation in the genomes of domestic animals.Anim Genet. 2012 Oct;43(5):503-17. doi: 10.1111/j.1365-2052.2012.02317.x. Epub 2012 Mar 6. Anim Genet. 2012. PMID: 22497594 Review.
Cited by
-
Genome-Wide Analysis Reveals Copy Number Variant Gene TGFBR3 Regulates Pig Back Fat Deposition.Animals (Basel). 2024 Sep 12;14(18):2657. doi: 10.3390/ani14182657. Animals (Basel). 2024. PMID: 39335247 Free PMC article.
-
c-di-GMP Regulates Various Phenotypes and Insecticidal Activity of Gram-Positive Bacillus thuringiensis.Front Microbiol. 2018 Feb 13;9:45. doi: 10.3389/fmicb.2018.00045. eCollection 2018. Front Microbiol. 2018. PMID: 29487570 Free PMC article.
-
Comprehensive Genome and Transcriptome Analysis Identifies SLCO3A1 Associated with Aggressive Behavior in Pigs.Biomolecules. 2023 Sep 12;13(9):1381. doi: 10.3390/biom13091381. Biomolecules. 2023. PMID: 37759782 Free PMC article.
-
Identification of copy number variations using high density whole-genome SNP markers in Chinese Dongxiang spotted pigs.Asian-Australas J Anim Sci. 2019 Dec 1;32(12):1809-1815. doi: 10.5713/ajas.18.0696. Epub 2019 Feb 7. Asian-Australas J Anim Sci. 2019. PMID: 30744341 Free PMC article.
-
CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole-genome sequencing.Gigascience. 2021 Nov 18;10(11):giab074. doi: 10.1093/gigascience/giab074. Gigascience. 2021. PMID: 34817058 Free PMC article.
References
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. - PubMed
-
- Li J, Jiang T, Mao JH, Balmain A, Peterson L, Harris C, et al. Genomic segmental polymorphisms in inbred mouse strains. Nat Genet. 2004;36(9):952–954. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

