Size-exclusion chromatography as a stand-alone methodology identifies novel markers in mass spectrometry analyses of plasma-derived vesicles from healthy individuals
- PMID: 26154623
- PMCID: PMC4495624
- DOI: 10.3402/jev.v4.27378
Size-exclusion chromatography as a stand-alone methodology identifies novel markers in mass spectrometry analyses of plasma-derived vesicles from healthy individuals
Abstract
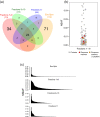
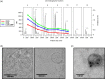
Plasma-derived vesicles hold a promising potential for use in biomedical applications. Two major challenges, however, hinder their implementation into translational tools: (a) the incomplete characterization of the protein composition of plasma-derived vesicles, in the size range of exosomes, as mass spectrometric analysis of plasma sub-components is recognizably troublesome and (b) the limited reach of vesicle-based studies in settings where the infrastructural demand of ultracentrifugation, the most widely used isolation/purification methodology, is not available. In this study, we have addressed both challenges by carrying-out mass spectrometry (MS) analyses of plasma-derived vesicles, in the size range of exosomes, from healthy donors obtained by 2 alternative methodologies: size-exclusion chromatography (SEC) on sepharose columns and Exo-Spin™. No exosome markers, as opposed to the most abundant plasma proteins, were detected by Exo-Spin™. In contrast, exosomal markers were present in the early fractions of SEC where the most abundant plasma proteins have been largely excluded. Noticeably, after a cross-comparative analysis of all published studies using MS to characterize plasma-derived exosomes from healthy individuals, we also observed a paucity of "classical exosome markers." Independent of the isolation method, however, we consistently identified 2 proteins, CD5 antigen-like (CD5L) and galectin-3-binding protein (LGALS3BP), whose presence was validated by a bead-exosome FACS assay. Altogether, our results support the use of SEC as a stand-alone methodology to obtain preparations of extracellular vesicles, in the size range of exosomes, from plasma and suggest the use of CD5L and LGALS3BP as more suitable markers of plasma-derived vesicles in MS.
Keywords: CD5L; LGALS3BP; comparative analysis; exosomes; low infrastructure settings; mass spectrometry; plasma-derived exosomes.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

