Identification of polarized macrophage subsets in zebrafish
- PMID: 26154973
- PMCID: PMC4521581
- DOI: 10.7554/eLife.07288
Identification of polarized macrophage subsets in zebrafish
Abstract
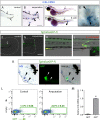
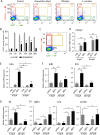
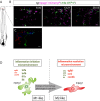
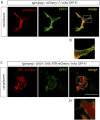
While the mammalian macrophage phenotypes have been intensively studied in vitro, the dynamic of their phenotypic polarization has never been investigated in live vertebrates. We used the zebrafish as a live model to identify and trail macrophage subtypes. We generated a transgenic line whose macrophages expressing tumour necrosis factor alpha (tnfa), a key feature of classically activated (M1) macrophages, express fluorescent proteins Tg(mpeg1:mCherryF/tnfa:eGFP-F). Using 4D-confocal microscopy, we showed that both aseptic wounding and Escherichia coli inoculation triggered macrophage recruitment, some of which started to express tnfa. RT-qPCR on Fluorescence Activated Cell Sorting (FACS)-sorted tnfa(+) and tnfa(-) macrophages showed that they, respectively, expressed M1 and alternatively activated (M2) mammalian markers. Fate tracing of tnfa(+) macrophages during the time-course of inflammation demonstrated that pro-inflammatory macrophages converted into M2-like phenotype during the resolution step. Our results reveal the diversity and plasticity of zebrafish macrophage subsets and underline the similarities with mammalian macrophages proposing a new system to study macrophage functional dynamic.
Keywords: developmental biology; immunology; live imaging; macrophages; stem cells; zebrafish.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



References
-
- Beider K, Bitner H, Leiba M, Gutwein O, Koren-Michowitz M, Ostrovsky O, Abraham M, Wald H, Galun E, Peled A, Nagler A. Multiple myeloma cells recruit tumor-supportive macrophages through the CXCR4/CXCL12 axis and promote their polarization toward the M2 phenotype. Oncotarget. 2014;5:11283–11296. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

