Metrics for the Human Proteome Project 2015: Progress on the Human Proteome and Guidelines for High-Confidence Protein Identification
- PMID: 26155816
- PMCID: PMC4755311
- DOI: 10.1021/acs.jproteome.5b00499
Metrics for the Human Proteome Project 2015: Progress on the Human Proteome and Guidelines for High-Confidence Protein Identification
Abstract
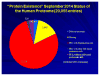
Remarkable progress continues on the annotation of the proteins identified in the Human Proteome and on finding credible proteomic evidence for the expression of "missing proteins". Missing proteins are those with no previous protein-level evidence or insufficient evidence to make a confident identification upon reanalysis in PeptideAtlas and curation in neXtProt. Enhanced with several major new data sets published in 2014, the human proteome presented as neXtProt, version 2014-09-19, has 16,491 unique confident proteins (PE level 1), up from 13,664 at 2012-12 and 15,646 at 2013-09. That leaves 2948 missing proteins from genes classified having protein existence level PE 2, 3, or 4, as well as 616 dubious proteins at PE 5. Here, we document the progress of the HPP and discuss the importance of assessing the quality of evidence, confirming automated findings and considering alternative protein matches for spectra and peptides. We provide guidelines for proteomics investigators to apply in reporting newly identified proteins.
Keywords: Global Proteome Machine database (GPMDB); HPP metrics; Human Protein Atlas; Human Proteome Project; PeptideAtlas; guidelines; high-confidence protein identifications; missing proteins; neXtProt; novel proteins.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
References
-
- Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, Deutsch EW, Domon B, Hancock W, He F, Hochstrasser D, Marko-Varga G, Salekdeh GH, Sechi S, Snyder M, Srivastava S, Uhlen M, Wu CH, Yamamoto T, Paik YK, Omenn GS. The human proteome project: current state and future direction. Mol Cell Proteomics. 2011;10(7):M111.009993. - PMC - PubMed
-
- Gaudet P, Argoud-Puy G, Cusin I, Duek P, Evalet O, Gateau A, Gleizes A, Pereira M, Zahn-Zabal M, Zwahlen C, Bairoch A, Lane L. neXtProt: organizing protein knowledge in the context of human proteome projects. J Proteome Res. 2013;12(1):293–8. - PubMed
-
- Marko-Varga G, Omenn GS, Paik YK, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J Proteome Res. 2013;12(1):1–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

