Proteomic analysis of Artemisia annua--towards elucidating the biosynthetic pathways of the antimalarial pro-drug artemisinin
- PMID: 26156581
- PMCID: PMC4496932
- DOI: 10.1186/s12870-015-0565-7
Proteomic analysis of Artemisia annua--towards elucidating the biosynthetic pathways of the antimalarial pro-drug artemisinin
Abstract
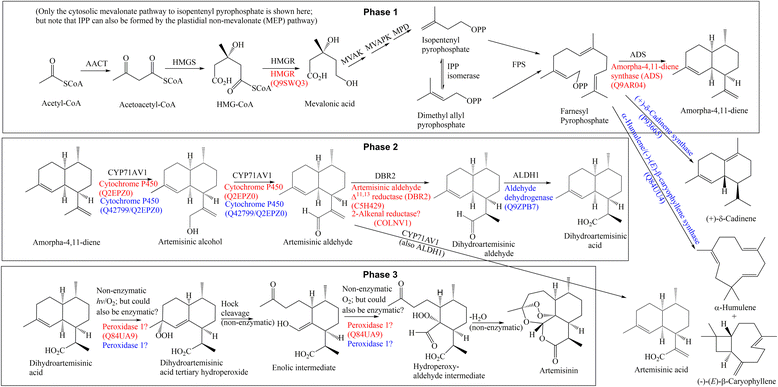
Background: MS-based proteomics was applied to the analysis of the medicinal plant Artemisia annua, exploiting a recently published contig sequence database (Graham et al. (2010) Science 327, 328-331) and other genomic and proteomic sequence databases for comparison. A. annua is the predominant natural source of artemisinin, the precursor for artemisinin-based combination therapies (ACTs), which are the WHO-recommended treatment for P. falciparum malaria.
Results: The comparison of various databases containing A. annua sequences (NCBInr/viridiplantae, UniProt/viridiplantae, UniProt/A. annua, an A. annua trichome Trinity contig database, the above contig database and another A. annua EST database) revealed significant differences in respect of their suitability for proteomic analysis, showing that an organism-specific database that has undergone extensive curation, leading to longer contig sequences, can greatly increase the number of true positive protein identifications, while reducing the number of false positives. Compared to previously published data an order-of-magnitude more proteins have been identified from trichome-enriched A. annua samples, including proteins which are known to be involved in the biosynthesis of artemisinin, as well as other highly abundant proteins, which suggest additional enzymatic processes occurring within the trichomes that are important for the biosynthesis of artemisinin.
Conclusions: The newly gained information allows for the possibility of an enzymatic pathway, utilizing peroxidases, for the less well understood final stages of artemisinin's biosynthesis, as an alternative to the known non-enzymatic in vitro conversion of dihydroartemisinic acid to artemisinin. Data are available via ProteomeXchange with identifier PXD000703.
Figures


Similar articles
-
TRICHOME AND ARTEMISININ REGULATOR 1 Is Required for Trichome Development and Artemisinin Biosynthesis in Artemisia annua.Mol Plant. 2015 Sep;8(9):1396-411. doi: 10.1016/j.molp.2015.04.002. Epub 2015 Apr 11. Mol Plant. 2015. PMID: 25870149
-
GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua.New Phytol. 2017 Apr;214(1):304-316. doi: 10.1111/nph.14373. Epub 2016 Dec 21. New Phytol. 2017. PMID: 28001315
-
The molecular cloning of dihydroartemisinic aldehyde reductase and its implication in artemisinin biosynthesis in Artemisia annua.Planta Med. 2010 Oct;76(15):1778-83. doi: 10.1055/s-0030-1249930. Epub 2010 May 19. Planta Med. 2010. PMID: 20486073
-
The biosynthesis of artemisinin (Qinghaosu) and the phytochemistry of Artemisia annua L. (Qinghao).Molecules. 2010 Oct 28;15(11):7603-98. doi: 10.3390/molecules15117603. Molecules. 2010. PMID: 21030913 Free PMC article. Review.
-
Transgenic approach to increase artemisinin content in Artemisia annua L.Plant Cell Rep. 2014 Apr;33(4):605-15. doi: 10.1007/s00299-014-1566-y. Epub 2014 Jan 12. Plant Cell Rep. 2014. PMID: 24413765 Review.
Cited by
-
Biotechnological approaches for artemisinin production in Artemisia.World J Microbiol Biotechnol. 2018 Mar 27;34(4):54. doi: 10.1007/s11274-018-2432-9. World J Microbiol Biotechnol. 2018. PMID: 29589124 Free PMC article. Review.
-
Recent Advances in the Synthetic Biology of Natural Drugs.Front Bioeng Biotechnol. 2021 Jul 29;9:691152. doi: 10.3389/fbioe.2021.691152. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34395399 Free PMC article. Review.
-
High variability of perezone content in rhizomes of Acourtia cordata wild plants, environmental factors related, and proteomic analysis.PeerJ. 2023 Nov 15;11:e16136. doi: 10.7717/peerj.16136. eCollection 2023. PeerJ. 2023. PMID: 38025722 Free PMC article.
-
Characterization of a class III peroxidase from Artemisia annua: relevance to artemisinin metabolism and beyond.Plant Mol Biol. 2019 Jul;100(4-5):527-541. doi: 10.1007/s11103-019-00879-x. Epub 2019 May 15. Plant Mol Biol. 2019. PMID: 31093899
-
Artemisia annua mutant impaired in artemisinin synthesis demonstrates importance of nonenzymatic conversion in terpenoid metabolism.Proc Natl Acad Sci U S A. 2016 Dec 27;113(52):15150-15155. doi: 10.1073/pnas.1611567113. Epub 2016 Dec 7. Proc Natl Acad Sci U S A. 2016. PMID: 27930305 Free PMC article.
References
-
- WHO: World Malaria Report 2014. 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

