REST represses a subset of the pancreatic endocrine differentiation program
- PMID: 26156633
- PMCID: PMC4732702
- DOI: 10.1016/j.ydbio.2015.07.002
REST represses a subset of the pancreatic endocrine differentiation program
Abstract
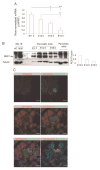
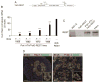
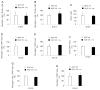
To contribute to devise successful beta-cell differentiation strategies for the cure of Type 1 diabetes we sought to uncover barriers that restrict endocrine fate acquisition by studying the role of the transcriptional repressor REST in the developing pancreas. Rest expression is prevented in neurons and in endocrine cells, which is necessary for their normal function. During development, REST represses a subset of genes in the neuronal differentiation program and Rest is down-regulated as neurons differentiate. Here, we investigate the role of REST in the differentiation of pancreatic endocrine cells, which are molecularly close to neurons. We show that Rest is widely expressed in pancreas progenitors and that it is down-regulated in differentiated endocrine cells. Sustained expression of REST in Pdx1(+) progenitors impairs the differentiation of endocrine-committed Neurog3(+) progenitors, decreases beta and alpha cell mass by E18.5, and triggers diabetes in adulthood. Conditional inactivation of Rest in Pdx1(+) progenitors is not sufficient to trigger endocrine differentiation but up-regulates a subset of differentiation genes. Our results show that the transcriptional repressor REST is active in pancreas progenitors where it gates the activation of part of the beta cell differentiation program.
Keywords: Beta cells; Diabetes; Islets; NRSF; Pancreas; Repressor.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



References
-
- Aoki H, Hara A, Era T, Kunisada T, Yamada Y. Genetic ablation of Rest leads to in vitro-specific derepression of neuronal genes during neurogenesis. Development. 2012;139:667–677. - PubMed
-
- Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. - PubMed
-
- Atouf F, Czernichow P, Scharfmann R. Expression of neuronal traits in pancreatic beta cells. Implication of neuron-restrictive silencing factor/re-pressor element silencing transcription factor, a neuron-restrictive silencer. J Biol Chem. 1997;272:1929–1934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

