Extra-Large G Proteins Expand the Repertoire of Subunits in Arabidopsis Heterotrimeric G Protein Signaling
- PMID: 26157115
- PMCID: PMC4577375
- DOI: 10.1104/pp.15.00251
Extra-Large G Proteins Expand the Repertoire of Subunits in Arabidopsis Heterotrimeric G Protein Signaling
Abstract
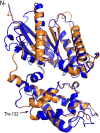
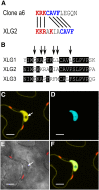
Heterotrimeric G proteins, consisting of Gα, Gβ, and Gγ subunits, are a conserved signal transduction mechanism in eukaryotes. However, G protein subunit numbers in diploid plant genomes are greatly reduced as compared with animals and do not correlate with the diversity of functions and phenotypes in which heterotrimeric G proteins have been implicated. In addition to GPA1, the sole canonical Arabidopsis (Arabidopsis thaliana) Gα subunit, Arabidopsis has three related proteins: the extra-large GTP-binding proteins XLG1, XLG2, and XLG3. We demonstrate that the XLGs can bind Gβγ dimers (AGB1 plus a Gγ subunit: AGG1, AGG2, or AGG3) with differing specificity in yeast (Saccharomyces cerevisiae) three-hybrid assays. Our in silico structural analysis shows that XLG3 aligns closely to the crystal structure of GPA1, and XLG3 also competes with GPA1 for Gβγ binding in yeast. We observed interaction of the XLGs with all three Gβγ dimers at the plasma membrane in planta by bimolecular fluorescence complementation. Bioinformatic and localization studies identified and confirmed nuclear localization signals in XLG2 and XLG3 and a nuclear export signal in XLG3, which may facilitate intracellular shuttling. We found that tunicamycin, salt, and glucose hypersensitivity and increased stomatal density are agb1-specific phenotypes that are not observed in gpa1 mutants but are recapitulated in xlg mutants. Thus, XLG-Gβγ heterotrimers provide additional signaling modalities for tuning plant G protein responses and increase the repertoire of G protein heterotrimer combinations from three to 12. The potential for signal partitioning and competition between the XLGs and GPA1 is a new paradigm for plant-specific cell signaling.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures









References
-
- Anderson DJ, Botella JR (2007) Expression analysis and subcellular localization of the Arabidopsis thaliana G-protein β-subunit AGB1. Plant Cell Rep 26: 1469–1480 - PubMed
-
- Bisht NC, Jez JM, Pandey S (2011) An elaborate heterotrimeric G-protein family from soybean expands the diversity of plant G-protein networks. New Phytol 190: 35–48 - PubMed
-
- Bitter GA, Egan KM (1984) Expression of heterologous genes in Saccharomyces cerevisiae from vectors utilizing the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Gene 32: 263–274 - PubMed
-
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

