Combinatorial synthesis and screening of cancer cell-specific nanomedicines targeted via phage fusion proteins
- PMID: 26157433
- PMCID: PMC4477153
- DOI: 10.3389/fmicb.2015.00628
Combinatorial synthesis and screening of cancer cell-specific nanomedicines targeted via phage fusion proteins
Abstract
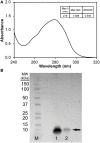
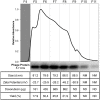
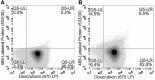
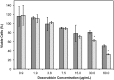
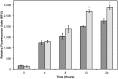
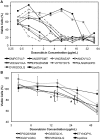
Active tumor targeting of nanomedicines has recently shown significant improvements in the therapeutic activity of currently existing drug delivery systems, such as liposomal doxorubicin (Doxil/Caelyx/Lipodox). Previously, we have shown that isolated pVIII major coat proteins of the fd-tet filamentous phage vector, containing cancer cell-specific peptide fusions at their N-terminus, can be used as active targeting ligands in a liposomal doxorubicin delivery system in vitro and in vivo. Here, we show a novel major coat protein isolation procedure in 2-propanol that allows spontaneous incorporation of the hydrophobic protein core into preformed liposomal doxorubicin with minimal damage or drug loss while still retaining the targeting ligand exposed for cell-specific targeting. Using a panel of 12 structurally unique ligands with specificity toward breast, lung, and/or pancreatic cancer, we showed the feasibility of pVIII major coat proteins to significantly increase the throughput of targeting ligand screening in a common nanomedicine core. Phage protein-modified Lipodox samples showed an average doxorubicin recovery of 82.8% across all samples with 100% of protein incorporation in the correct orientation (N-terminus exposed). Following cytotoxicity screening in a doxorubicin-sensitive breast cancer line (MCF-7), three major groups of ligands were identified. Ligands showing the most improved cytotoxicity included: DMPGTVLP, ANGRPSMT, VNGRAEAP, and ANDVYLD showing a 25-fold improvement (p < 0.05) in toxicity. Similarly DGQYLGSQ, ETYNQPYL, and GSSEQLYL ligands with specificity toward a doxorubicin-insensitive pancreatic cancer line (PANC-1) showed significant increases in toxicity (2-fold; p < 0.05). Thus, we demonstrated proof-of-concept that pVIII major coat proteins can be screened in significantly higher throughput to identify novel ligands displaying improved therapeutic activity in a desired cancer phenotype.
Keywords: breast cancer; doxorubicin; liposomal drug delivery; pancreatic cancer; phage display; targeted nanomedicines.
Figures


Similar articles
-
Selection of pancreatic cancer cell-binding landscape phages and their use in development of anticancer nanomedicines.Protein Eng Des Sel. 2014 Jul;27(7):235-43. doi: 10.1093/protein/gzu020. Epub 2014 Jun 4. Protein Eng Des Sel. 2014. PMID: 24899628 Free PMC article.
-
Enhanced binding and killing of target tumor cells by drug-loaded liposomes modified with tumor-specific phage fusion coat protein.Nanomedicine (Lond). 2010 Jun;5(4):563-74. doi: 10.2217/nnm.10.30. Nanomedicine (Lond). 2010. PMID: 20528452 Free PMC article.
-
Enhanced tumor delivery and antitumor activity in vivo of liposomal doxorubicin modified with MCF-7-specific phage fusion protein.Nanomedicine. 2014 Feb;10(2):421-30. doi: 10.1016/j.nano.2013.08.009. Epub 2013 Sep 9. Nanomedicine. 2014. PMID: 24028893 Free PMC article.
-
Ligand-targeted particulate nanomedicines undergoing clinical evaluation: current status.Adv Drug Deliv Rev. 2013 Oct;65(10):1284-98. doi: 10.1016/j.addr.2013.08.012. Epub 2013 Sep 6. Adv Drug Deliv Rev. 2013. PMID: 24018362 Review.
-
Phage protein-targeted cancer nanomedicines.FEBS Lett. 2014 Jan 21;588(2):341-9. doi: 10.1016/j.febslet.2013.11.011. Epub 2013 Nov 20. FEBS Lett. 2014. PMID: 24269681 Free PMC article. Review.
Cited by
-
Mimotopes for Mycotoxins Diagnosis Based on Random Peptides or Recombinant Antibodies from Phage Library.Molecules. 2021 Dec 17;26(24):7652. doi: 10.3390/molecules26247652. Molecules. 2021. PMID: 34946736 Free PMC article. Review.
-
Selection of Lung Cancer-Specific Landscape Phage for Targeted Drug Delivery.Comb Chem High Throughput Screen. 2016;19(5):412-22. doi: 10.2174/1386207319666160420141024. Comb Chem High Throughput Screen. 2016. PMID: 27095536 Free PMC article.
-
Promiscuous tumor targeting phage proteins.Protein Eng Des Sel. 2016 Mar;29(3):93-103. doi: 10.1093/protein/gzv064. Epub 2016 Jan 12. Protein Eng Des Sel. 2016. PMID: 26764410 Free PMC article.
-
Phage-Displayed Mimotopes of SARS-CoV-2 Spike Protein Targeted to Authentic and Alternative Cellular Receptors.Viruses. 2022 Feb 14;14(2):384. doi: 10.3390/v14020384. Viruses. 2022. PMID: 35215976 Free PMC article.
-
Detection of cancer stem cells by EMT-specific biomarker-based peptide ligands.Sci Rep. 2021 Nov 17;11(1):22430. doi: 10.1038/s41598-021-01138-0. Sci Rep. 2021. PMID: 34789743 Free PMC article.
References
-
- Brigati J. R., Samoylova T. I., Jayanna P. K., Petrenko V. A. (2008). Phage display for generating peptide reagents, in Current Protocols in Protein Science, eds Coligan J. E., Dunn B. M., Speicher D. W., Wingfield P. T. (New Jersey, NJ: John Wiley & Sons, Inc; ), 1–27. 10.1002/0471140864.ps1809s51 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

