TGFβ-Mediated induction of SphK1 as a potential determinant in human MDA-MB-231 breast cancer cell bone metastasis
- PMID: 26157579
- PMCID: PMC4495778
- DOI: 10.1038/bonekey.2015.88
TGFβ-Mediated induction of SphK1 as a potential determinant in human MDA-MB-231 breast cancer cell bone metastasis
Abstract
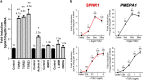
Mechanistic understanding of the preferential homing of circulating tumor cells to bone and their perturbation on bone metabolism within the tumor-bone microenvironment remains poorly understood. Alteration in both transforming growth factor β (TGFβ) signaling and sphingolipid metabolism results in the promotion of tumor growth and metastasis. Previous studies using MDA-MB-231 human breast cancer-derived cell lines of variable metastatic potential were queried for changes in sphingolipid metabolism genes to explore correlations between TGFβ dependence and bone metastatic behavior. Of these genes, only sphingosine kinase-1 (SPHK1) was identified to be significantly increased following TGFβ treatment. Induction of SPHK1 expression correlated to the degree of metastatic capacity in these MDA-MB-231-derived cell lines. We demonstrate that TGFβ mediates the regulation of SPHK1 gene expression, protein kinase activity and is critical to MDA-MB-231 cell viability. Furthermore, a bioinformatic analysis of human breast cancer gene expression supports SPHK1 as a hallmark TGFβ target gene that also bears the genetic fingerprint of the basal-like/triple-negative breast cancer molecular subtype. These data suggest a potential new signaling axis between TGFβ/SphK1 that may have a role in the development, prognosis or the clinical phenotype associated with tumor-bone metastasis.
Conflict of interest statement
TAG received commercial research grants from AstraZeneca and Exelexis and is a consultant/advisory board member of Novartis. KRS, JKM, DFE and HHB are employees of Eli Lilly and Company. The remaining authors declare no conflict of interest.
Figures






Similar articles
-
Metastatic triple-negative breast cancer is dependent on SphKs/S1P signaling for growth and survival.Cell Signal. 2017 Apr;32:85-92. doi: 10.1016/j.cellsig.2017.01.021. Epub 2017 Jan 17. Cell Signal. 2017. PMID: 28108260 Free PMC article.
-
Predictive and prognostic value of sphingosine kinase 1 expression in patients with invasive ductal carcinoma of the breast.Am J Transl Res. 2017 Dec 15;9(12):5684-5695. eCollection 2017. Am J Transl Res. 2017. PMID: 29312521 Free PMC article.
-
Triple Negative Breast Cancer Depends on Sphingosine Kinase 1 (SphK1)/Sphingosine-1-Phosphate (S1P)/Sphingosine 1-Phosphate Receptor 3 (S1PR3)/Notch Signaling for Metastasis.Med Sci Monit. 2018 Apr 1;24:1912-1923. doi: 10.12659/msm.905833. Med Sci Monit. 2018. PMID: 29605826 Free PMC article.
-
Parathyroid hormone-related protein and bone metastases.Cancer. 1997 Oct 15;80(8 Suppl):1572-80. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1572::aid-cncr7>3.3.co;2-d. Cancer. 1997. PMID: 9362424 Review.
-
Molecular mechanisms of osteolytic bone metastases.Cancer. 2000 Jun 15;88(12 Suppl):2892-8. doi: 10.1002/1097-0142(20000615)88:12+<2892::aid-cncr2>3.0.co;2-y. Cancer. 2000. PMID: 10898330 Review.
Cited by
-
miR-659-3p is involved in the regulation of the chemotherapy response of colorectal cancer via modulating the expression of SPHK1.Am J Cancer Res. 2016 Sep 1;6(9):1976-1985. eCollection 2016. Am J Cancer Res. 2016. PMID: 27725903 Free PMC article.
-
Signaling Crosstalk of TGF-β/ALK5 and PAR2/PAR1: A Complex Regulatory Network Controlling Fibrosis and Cancer.Int J Mol Sci. 2018 May 24;19(6):1568. doi: 10.3390/ijms19061568. Int J Mol Sci. 2018. PMID: 29795022 Free PMC article. Review.
-
A novel sphingosine kinase 1 inhibitor (SKI-5C) induces cell death of Wilms' tumor cells in vitro and in vivo.Am J Transl Res. 2016 Nov 15;8(11):4548-4563. eCollection 2016. Am J Transl Res. 2016. PMID: 27904661 Free PMC article.
-
Biglycan- and Sphingosine Kinase-1 Signaling Crosstalk Regulates the Synthesis of Macrophage Chemoattractants.Int J Mol Sci. 2017 Mar 9;18(3):595. doi: 10.3390/ijms18030595. Int J Mol Sci. 2017. PMID: 28282921 Free PMC article.
-
Pro-Survival Lipid Sphingosine-1-Phosphate Metabolically Programs T Cells to Limit Anti-tumor Activity.Cell Rep. 2019 Aug 13;28(7):1879-1893.e7. doi: 10.1016/j.celrep.2019.07.044. Cell Rep. 2019. PMID: 31412253 Free PMC article.
References
-
- Kok JW, Sietsma H. Sphingolipid metabolism enzymes as targets for anticancer therapy. Curr Drug Targets 2004; 5: 375–382. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
