Global sensitivity analysis of a dynamic model for gene expression in Drosophila embryos
- PMID: 26157608
- PMCID: PMC4476099
- DOI: 10.7717/peerj.1022
Global sensitivity analysis of a dynamic model for gene expression in Drosophila embryos
Abstract
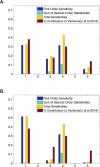
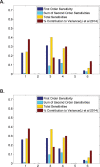
It is well known that gene regulation is a tightly controlled process in early organismal development. However, the roles of key processes involved in this regulation, such as transcription and translation, are less well understood, and mathematical modeling approaches in this field are still in their infancy. In recent studies, biologists have taken precise measurements of protein and mRNA abundance to determine the relative contributions of key factors involved in regulating protein levels in mammalian cells. We now approach this question from a mathematical modeling perspective. In this study, we use a simple dynamic mathematical model that incorporates terms representing transcription, translation, mRNA and protein decay, and diffusion in an early Drosophila embryo. We perform global sensitivity analyses on this model using various different initial conditions and spatial and temporal outputs. Our results indicate that transcription and translation are often the key parameters to determine protein abundance. This observation is in close agreement with the experimental results from mammalian cells for various initial conditions at particular time points, suggesting that a simple dynamic model can capture the qualitative behavior of a gene. Additionally, we find that parameter sensitivites are temporally dynamic, illustrating the importance of conducting a thorough global sensitivity analysis across multiple time points when analyzing mathematical models of gene regulation.
Keywords: Gene expression; Parameter sensitivity; Reaction–diffusion; Transcription; Translation.
Conflict of interest statement
The authors declare that there are no competing interests.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

