Structural modeling of the N-terminal signal-receiving domain of IκBα
- PMID: 26157801
- PMCID: PMC4477481
- DOI: 10.3389/fmolb.2015.00032
Structural modeling of the N-terminal signal-receiving domain of IκBα
Abstract
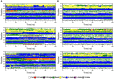
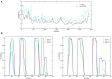
The transcription factor nuclear factor-κB (NF-κB) exerts essential roles in many biological processes including cell growth, apoptosis and innate and adaptive immunity. The NF-κB inhibitor (IκBα) retains NF-κB in the cytoplasm and thus inhibits nuclear localization of NF-κB and its association with DNA. Recent protein crystal structures of the C-terminal part of IκBα in complex with NF-κB provided insights into the protein-protein interactions but could not reveal structural details about the N-terminal signal receiving domain (SRD). The SRD of IκBα contains a degron, formed following phosphorylation by IκB kinases (IKK). In current protein X-ray structures, however, the SRD is not resolved and assumed to be disordered. Here, we combined secondary structure annotation and domain threading followed by long molecular dynamics (MD) simulations and showed that the SRD possesses well-defined secondary structure elements. We show that the SRD contains 3 additional stable α-helices supplementing the six ARDs present in crystallized IκBα. The IκBα/NF-κB protein-protein complex remained intact and stable during the entire simulations. Also in solution, free IκBα retains its structural integrity. Differences in structural topology and dynamics were observed by comparing the structures of NF-κB free and NF-κB bound IκBα-complex. This study paves the way for investigating the signaling properties of the SRD in the IκBα degron. A detailed atomic scale understanding of molecular mechanism of NF-κB activation, regulation and the protein-protein interactions may assist to design and develop novel chronic inflammation modulators.
Keywords: IκBα; N-terminal extension; NF-κB; molecular dynamics simulation; protein-protein complex refinement; secondary structure prediction; signal receiving domain; signal transduction.
Figures








References
LinkOut - more resources
Full Text Sources
Other Literature Sources

