A palette of fluorescent proteins optimized for diverse cellular environments
- PMID: 26158227
- PMCID: PMC4499870
- DOI: 10.1038/ncomms8670
A palette of fluorescent proteins optimized for diverse cellular environments
Abstract
To perform quantitative live cell imaging, investigators require fluorescent reporters that accurately report protein localization and levels, while minimally perturbing the cell. Yet, within the biochemically distinct environments of cellular organelles, popular fluorescent proteins (FPs), including EGFP, can be unreliable for quantitative imaging, resulting in the underestimation of protein levels and incorrect localization. Specifically, within the secretory pathway, significant populations of FPs misfold and fail to fluoresce due to non-native disulphide bond formation. Furthermore, transmembrane FP-fusion constructs can disrupt organelle architecture due to oligomerizing tendencies of numerous common FPs. Here, we describe a powerful set of bright and inert FPs optimized for use in multiple cellular compartments, especially oxidizing environments and biological membranes. Also, we provide new insights into the use of red FPs in the secretory pathway. Our monomeric 'oxFPs' finally resolve long-standing, underappreciated and important problems of cell biology and should be useful for a number of applications.
Conflict of interest statement
A provisional patent application has been filed covering part of the results described in this manuscript (L.M.C. and E.L.S). Albert Einstein College Medicine and E.L.S. have licensed technology described in this manuscript to Lucigen Corp. The remaining authors declare no competing financial interests.
Figures

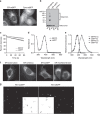
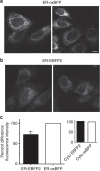

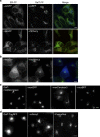

References
-
- Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. - PubMed
-
- Rizzo MA, Davidson MW, Piston DW. Fluorescent protein tracking and detection: fluorescent protein structure and color variants. Cold Spring Harb. Protoc. 2009;2009:pdb.top63. - PubMed
-
- Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J. Cell Sci. 2007;120:4247–4260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

