PTEN regulates DNA replication progression and stalled fork recovery
- PMID: 26158445
- PMCID: PMC4499867
- DOI: 10.1038/ncomms8620
PTEN regulates DNA replication progression and stalled fork recovery
Abstract
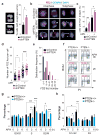
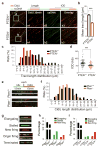
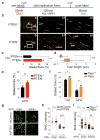
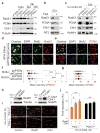
Faithful DNA replication is a cornerstone of genomic integrity. PTEN plays multiple roles in genome protection and tumour suppression. Here we report on the importance of PTEN in DNA replication. PTEN depletion leads to impairment of replication progression and stalled fork recovery, indicating an elevation of endogenous replication stress. Exogenous replication inhibition aggravates replication-originated DNA lesions without inducing S phase arrest in cells lacking PTEN, representing replication stress tolerance. iPOND analysis reveals the physical association of PTEN with DNA replication forks and PTEN-dependent recruitment of Rad51. PTEN deletion results in Rad51 dissociation from replication forks. Stalled replication forks in Pten-null cells can be reactivated by ectopic Rad51 or PTEN, the latter facilitating chromatin loading of Rad51. These data highlight the interplay of PTEN with Rad51 in promoting stalled fork restart. We propose that loss of PTEN may initiate a replication stress cascade that progressively deteriorates through the cell cycle.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

