CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination
- PMID: 26158450
- PMCID: PMC4499871
- DOI: 10.1038/ncomms8676
CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination
Abstract
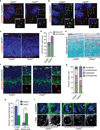
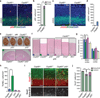
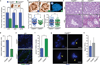
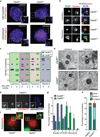
CEP63 is a centrosomal protein that facilitates centriole duplication and is regulated by the DNA damage response. Mutations in CEP63 cause Seckel syndrome, a human disease characterized by microcephaly and dwarfism. Here we demonstrate that Cep63-deficient mice recapitulate Seckel syndrome pathology. The attrition of neural progenitor cells involves p53-dependent cell death, and brain size is rescued by the deletion of p53. Cell death is not the result of an aberrant DNA damage response but is triggered by centrosome-based mitotic errors. In addition, Cep63 loss severely impairs meiotic recombination, leading to profound male infertility. Cep63-deficient spermatocytes display numerical and structural centrosome aberrations, chromosome entanglements and defective telomere clustering, suggesting that a reduction in centrosome-mediated chromosome movements underlies recombination failure. Our results provide novel insight into the molecular pathology of microcephaly and establish a role for the centrosome in meiotic recombination.
Figures



References
-
- O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

