The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in Streptococcus mutans
- PMID: 26158727
- PMCID: PMC4497675
- DOI: 10.1371/journal.pgen.1005353
The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in Streptococcus mutans
Abstract
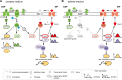
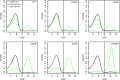
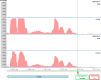
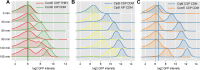
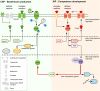
Two small quorum sensing (QS) peptides regulate competence in S. mutans in a cell density dependent manner: XIP (sigX inducing peptide) and CSP (competence stimulating peptide). Depending on the environmental conditions isogenic S. mutans cells can split into a competent and non-competent subpopulation. The origin of this population heterogeneity has not been experimentally determined and it is unknown how the two QS systems are connected. We developed a toolbox of single and dual fluorescent reporter strains and systematically knocked out key genes of the competence signaling cascade in the reporter strain backgrounds. By following signal propagation on the single cell level we discovered that the master regulator of competence, the alternative sigma factor SigX, directly controls expression of the response regulator for bacteriocin synthesis ComE. Consequently, a SigX binding motif (cin-box) was identified in the promoter region of comE. Overexpressing the genetic components involved in competence development demonstrated that ComRS represents the origin of bimodality and determines the modality of the downstream regulators SigX and ComE. Moreover these analysis showed that there is no direct regulatory link between the two QS signaling cascades. Competence is induced through a hierarchical XIP signaling cascade, which has no regulatory input from the CSP cascade. CSP exclusively regulates bacteriocin synthesis. We suggest renaming it mutacin inducing peptide (MIP). Finally, using phosphomimetic comE mutants we show that unimodal bacteriocin production is controlled posttranslationally, thus solving the puzzling observation that in complex media competence is observed in a subpopulation only, while at the same time all cells produce bacteriocins. The control of both bacteriocin synthesis and competence through the alternative sigma-factor SigX suggests that S. mutans increases its genetic repertoire via QS controlled predation on neighboring species in its natural habitat.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3: 711–721. nrmicro1234 [pii]. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

