Functional Analyses of Resurrected and Contemporary Enzymes Illuminate an Evolutionary Path for the Emergence of Exolysis in Polysaccharide Lyase Family 2
- PMID: 26160170
- PMCID: PMC4571855
- DOI: 10.1074/jbc.M115.664847
Functional Analyses of Resurrected and Contemporary Enzymes Illuminate an Evolutionary Path for the Emergence of Exolysis in Polysaccharide Lyase Family 2
Abstract
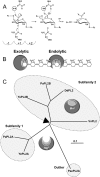
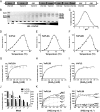
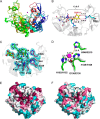
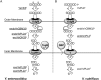
Family 2 polysaccharide lyases (PL2s) preferentially catalyze the β-elimination of homogalacturonan using transition metals as catalytic cofactors. PL2 is divided into two subfamilies that have been generally associated with secretion, Mg(2+) dependence, and endolysis (subfamily 1) and with intracellular localization, Mn(2+) dependence, and exolysis (subfamily 2). When present within a genome, PL2 genes are typically found as tandem copies, which suggests that they provide complementary activities at different stages along a catabolic cascade. This relationship most likely evolved by gene duplication and functional divergence (i.e. neofunctionalization). Although the molecular basis of subfamily 1 endolytic activity is understood, the adaptations within the active site of subfamily 2 enzymes that contribute to exolysis have not been determined. In order to investigate this relationship, we have conducted a comparative enzymatic analysis of enzymes dispersed within the PL2 phylogenetic tree and elucidated the structure of VvPL2 from Vibrio vulnificus YJ016, which represents a transitional member between subfamiles 1 and 2. In addition, we have used ancestral sequence reconstruction to functionally investigate the segregated evolutionary history of PL2 progenitor enzymes and illuminate the molecular evolution of exolysis. This study highlights that ancestral sequence reconstruction in combination with the comparative analysis of contemporary and resurrected enzymes holds promise for elucidating the origins and activities of other carbohydrate active enzyme families and the biological significance of cryptic metabolic pathways, such as pectinolysis within the zoonotic marine pathogen V. vulnificus.
Keywords: enzyme kinetics; enzyme structure; exolysis; magnesium; manganese; polysaccharide lyase; protein evolution.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Lombard V., Bernard T., Rancurel C., Brumer H., Coutinho P. M., Henrissat B. (2010) A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444 - PubMed
-
- Abbott D. W., Boraston A. B. (2007) A family 2 pectate lyase displays a rare fold and transition metal-assisted β-elimination. J. Biol. Chem. 282, 35328–35336 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

