Dimerization Mediated by a Divergent Forkhead-associated Domain Is Essential for the DNA Damage and Spindle Functions of Fission Yeast Mdb1
- PMID: 26160178
- PMCID: PMC4543663
- DOI: 10.1074/jbc.M115.642538
Dimerization Mediated by a Divergent Forkhead-associated Domain Is Essential for the DNA Damage and Spindle Functions of Fission Yeast Mdb1
Abstract
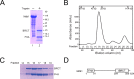
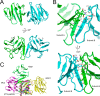
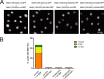
MDC1 is a key factor of DNA damage response in mammalian cells. It possesses two phospho-binding domains. In its C terminus, a tandem BRCA1 C-terminal domain binds phosphorylated histone H2AX, and in its N terminus, a forkhead-associated (FHA) domain mediates a phosphorylation-enhanced homodimerization. The FHA domain of the Drosophila homolog of MDC1, MU2, also forms a homodimer but utilizes a different dimer interface. The functional importance of the dimerization of MDC1 family proteins is uncertain. In the fission yeast Schizosaccharomyces pombe, a protein sharing homology with MDC1 in the tandem BRCA1 C-terminal domain, Mdb1, regulates DNA damage response and mitotic spindle functions. Here, we report the crystal structure of the N-terminal 91 amino acids of Mdb1. Despite a lack of obvious sequence conservation to the FHA domain of MDC1, this region of Mdb1 adopts an FHA-like fold and is therefore termed Mdb1-FHA. Unlike canonical FHA domains, Mdb1-FHA lacks all the conserved phospho-binding residues. It forms a stable homodimer through an interface distinct from those of MDC1 and MU2. Mdb1-FHA is important for the localization of Mdb1 to DNA damage sites and the spindle midzone, contributes to the roles of Mdb1 in cellular responses to genotoxins and an antimicrotubule drug, and promotes in vitro binding of Mdb1 to a phospho-H2A peptide. The defects caused by the loss of Mdb1-FHA can be rescued by fusion with either of two heterologous dimerization domains, suggesting that the main function of Mdb1-FHA is mediating dimerization. Our data support that FHA-mediated dimerization is conserved for MDC1 family proteins.
Keywords: DNA damage response; H2A histone family, member X (H2AFX); MDC1; Mdb1; Schizosaccharomyces pombe; crystal structure; dimerization; forkhead-associated domain (FHA domain); mitotic spindle; protein-protein interaction.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Reinhardt H. C., Yaffe M. B. (2013) Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14, 563–580 - PubMed
-
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 - PubMed
-
- Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 123, 1213–1226 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

