Redox Activation of the Universally Conserved ATPase YchF by Thioredoxin 1
- PMID: 26160547
- PMCID: PMC4742990
- DOI: 10.1089/ars.2015.6272
Redox Activation of the Universally Conserved ATPase YchF by Thioredoxin 1
Abstract
Aims: YchF/Ola1 are unconventional members of the universally conserved GTPase family because they preferentially hydrolyze ATP rather than GTP. These ATPases have been associated with various cellular processes and pathologies, including DNA repair, tumorigenesis, and apoptosis. In particular, a possible role in regulating the oxidative stress response has been suggested for both bacterial and human YchF/Ola1. In this study, we analyzed how YchF responds to oxidative stress and how it potentially regulates the antioxidant response.
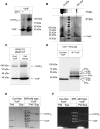
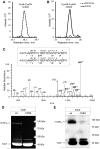
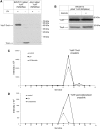
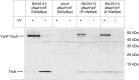
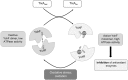
Results: Our data identify a redox-regulated monomer-dimer equilibrium of YchF as a key event in the functional cycle of YchF. Upon oxidative stress, the oxidation of a conserved and surface-exposed cysteine residue promotes YchF dimerization, which is accompanied by inhibition of the ATPase activity. No dimers were observed in a YchF mutant lacking this cysteine. In vitro, the YchF dimer is dissociated by thioredoxin 1 (TrxA) and this stimulates the ATPase activity. The physiological significance of the YchF-thioredoxin 1 interaction was demonstrated by in vivo cross-linking, which validated this interaction in living cells. This approach also revealed that both the ATPase domain and the helical domain of YchF are in contact with TrxA.
Innovation: YchF/Ola1 are the first redox-regulated members of the universally conserved GTPase family and are inactivated by oxidation of a conserved cysteine residue within the nucleotide-binding motif.
Conclusion: Our data provide novel insights into the regulation of the so far ill-defined YchF/Ola1 family of proteins and stipulate their role as negative regulators of the oxidative stress response.
Figures



References
-
- Akhova AV. and Tkachenko AG. ATP/ADP alteration as a sign of the oxidative stress development in Escherichia coli under antibiotic treatment. FEMS Microbiol Lett 353: 69–76, 2014 - PubMed
-
- Alexander RW. and Schimmel P. Domain-domain communication in aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol 69: 317–349, 2001 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
