Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage
- PMID: 26160905
- PMCID: PMC4550963
- DOI: 10.1242/dev.119339
Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage
Abstract
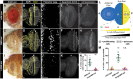
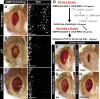
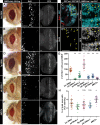
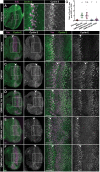
Regeneration of damaged tissues typically requires a population of active stem cells. How damaged tissue is regenerated in quiescent tissues lacking a stem cell population is less well understood. We used a genetic screen in the developing Drosophila melanogaster eye to investigate the mechanisms that trigger quiescent cells to re-enter the cell cycle and proliferate in response to tissue damage. We discovered that Hippo signaling regulates compensatory proliferation after extensive cell death in the developing eye. Scalloped and Yorkie, transcriptional effectors of the Hippo pathway, drive Cyclin E expression to induce cell cycle re-entry in cells that normally remain quiescent in the absence of damage. Ajuba, an upstream regulator of Hippo signaling that functions as a sensor of epithelial integrity, is also required for cell cycle re-entry. Thus, in addition to its well-established role in modulating proliferation during periods of tissue growth, Hippo signaling maintains homeostasis by regulating quiescent cell populations affected by tissue damage.
Keywords: Apoptosis; Cell cycle; Compensatory proliferation; Drosophila; Hippo signaling; Quiescence; Regeneration.
© 2015. Published by The Company of Biologists Ltd.
Figures
Similar articles
-
Loss of the Drosophila cell polarity regulator Scribbled promotes epithelial tissue overgrowth and cooperation with oncogenic Ras-Raf through impaired Hippo pathway signaling.BMC Dev Biol. 2011 Sep 29;11:57. doi: 10.1186/1471-213X-11-57. BMC Dev Biol. 2011. PMID: 21955824 Free PMC article.
-
Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador-Warts-Hippo pathway.Cell Death Differ. 2011 Aug;18(8):1346-55. doi: 10.1038/cdd.2011.6. Epub 2011 Feb 11. Cell Death Differ. 2011. PMID: 21311569 Free PMC article.
-
Validating upstream regulators of Yorkie activity in Hippo signaling through scalloped-based genetic epistasis.Development. 2018 Feb 21;145(4):dev157545. doi: 10.1242/dev.157545. Development. 2018. PMID: 29467233 Free PMC article.
-
A role for Hipk in the Hippo pathway.Sci Signal. 2013 May 14;6(275):pe18. doi: 10.1126/scisignal.2004259. Sci Signal. 2013. PMID: 23674821 Review.
-
Hippo signaling in Drosophila: recent advances and insights.Dev Dyn. 2012 Jan;241(1):3-15. doi: 10.1002/dvdy.22723. Epub 2011 Aug 25. Dev Dyn. 2012. PMID: 22174083 Free PMC article. Review.
Cited by
-
Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease.Cell Death Differ. 2017 Aug;24(8):1390-1400. doi: 10.1038/cdd.2017.47. Epub 2017 Mar 31. Cell Death Differ. 2017. PMID: 28362431 Free PMC article. Review.
-
Fate mapping during regeneration: Cells that undergo compensatory proliferation in damaged Drosophila eye imaginal discs differentiate into multiple retinal accessory cell types.Dev Biol. 2018 Dec 15;444(2):43-49. doi: 10.1016/j.ydbio.2018.10.011. Epub 2018 Oct 19. Dev Biol. 2018. PMID: 30347187 Free PMC article.
-
Two Sides of the Same Coin - Compensatory Proliferation in Regeneration and Cancer.Adv Exp Med Biol. 2019;1167:65-85. doi: 10.1007/978-3-030-23629-8_4. Adv Exp Med Biol. 2019. PMID: 31520349 Review.
-
Imaginal Disc Regeneration: Something Old, Something New.Cold Spring Harb Perspect Biol. 2022 Nov 1;14(11):a040733. doi: 10.1101/cshperspect.a040733. Cold Spring Harb Perspect Biol. 2022. PMID: 34872971 Free PMC article. Review.
-
Upstream paths for Hippo signaling in Drosophila organ development.BMB Rep. 2018 Mar;51(3):134-142. doi: 10.5483/bmbrep.2018.51.3.027. BMB Rep. 2018. PMID: 29397870 Free PMC article. Review.
References
-
- Bedelbaeva K., Snyder A., Gourevitch D., Clark L., Zhang X.-M., Leferovich J., Cheverud J. M., Lieberman P. and Heber-Katz E. (2010). Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc. Natl. Acad. Sci. USA 107, 5845-5850. 10.1073/pnas.1000830107 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

