Genetics of human metabolism: an update
- PMID: 26160913
- PMCID: PMC4572003
- DOI: 10.1093/hmg/ddv263
Genetics of human metabolism: an update
Abstract
Genome-wide association studies with metabolomics (mGWAS) identify genetically influenced metabotypes (GIMs), their ensemble defining the heritable part of every human's metabolic individuality. Knowledge of genetic variation in metabolism has many applications of biomedical and pharmaceutical interests, including the functional understanding of genetic associations with clinical end points, design of strategies to correct dysregulations in metabolic disorders and the identification of genetic effect modifiers of metabolic disease biomarkers. Furthermore, it has been shown that GIMs provide testable hypotheses for functional genomics and metabolomics and for the identification of novel gene functions and metabolite identities. mGWAS with growing sample sizes and increasingly complex metabolic trait panels are being conducted, allowing for more comprehensive and systems-based downstream analyses. The generated large datasets of genetic associations can now be mined by the biomedical research community and provide valuable resources for hypothesis-driven studies. In this review, we provide a brief summary of the key aspects of mGWAS, followed by an update of recently published mGWAS. We then discuss new approaches of integrating and exploring mGWAS results and finish by presenting selected applications of GIMs in recent studies.
© The Author 2015. Published by Oxford University Press.
Figures

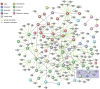
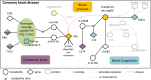
References
-
- Nicholson J.K., Lindon J.C. (2008) Systems biology: metabonomics. Nature, 455, 1054–1056. - PubMed
-
- Suhre K., Gieger C. (2012) Genetic variation in metabolic phenotypes: study designs and applications. Nat. Rev. Genet., 13, 759–769. - PubMed
-
- Gieger C., Geistlinger L., Altmaier E., Hrabe de Angelis M., Kronenberg F., Meitinger T., Mewes H.W., Wichmann H.E., Weinberger K.M., Adamski J. et al. (2008) Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS. Genet., 4, e1000282. - PMC - PubMed
-
- Kronenberg F. (2012) Metabolic traits as intermediate phenotypes. In Suhre K. (ed), Genetics Meets Metabolomics. Springer, New York, pp. 255–264.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

