Rhizobium-legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS
- PMID: 26161635
- PMCID: PMC4681849
- DOI: 10.1038/ismej.2015.103
Rhizobium-legume symbiosis in the absence of Nod factors: two possible scenarios with or without the T3SS
Abstract
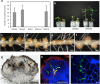
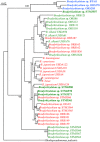
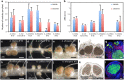
The occurrence of alternative Nod factor (NF)-independent symbiosis between legumes and rhizobia was first demonstrated in some Aeschynomene species that are nodulated by photosynthetic bradyrhizobia lacking the canonical nodABC genes. In this study, we revealed that a large diversity of non-photosynthetic bradyrhizobia, including B. elkanii, was also able to induce nodules on the NF-independent Aeschynomene species, A. indica. Using cytological analysis of the nodules and the nitrogenase enzyme activity as markers, a gradient in the symbiotic interaction between bradyrhizobial strains and A. indica could be distinguished. This ranged from strains that induced nodules that were only infected intercellularly to rhizobial strains that formed nodules in which the host cells were invaded intracellularly and that displayed a weak nitrogenase activity. In all non-photosynthetic bradyrhizobia, the type III secretion system (T3SS) appears required to trigger nodule organogenesis. In contrast, genome sequence analysis revealed that apart from a few exceptions, like the Bradyrhizobium ORS285 strain, photosynthetic bradyrhizobia strains lack a T3SS. Furthermore, analysis of the symbiotic properties of an ORS285 T3SS mutant revealed that the T3SS could have a positive or negative role for the interaction with NF-dependent Aeschynomene species, but that it is dispensable for the interaction with all NF-independent Aeschynomene species tested. Taken together, these data indicate that two NF-independent symbiotic processes are possible between legumes and rhizobia: one dependent on a T3SS and one using a so far unknown mechanism.
Figures

References
-
- Aslam SN, Newman MA, Erbs G, Morrissey KL, Chinchilla D, Boller T et al. (2008). Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr Biol 18: 1078–1083. - PubMed
-
- Bonaldi K, Gargani D, Prin Y, Fardoux J, Gully D, Nouwen N et al. (2011). Nodulation of Aeschynomeneafraspera and A. indica by photosynthetic Bradyrhizobium sp. strain ORS285: the Nod-dependent versus the Nod-independent symbiotic interaction. Mol Plant Microbe Interact 24: 1359–1371. - PubMed
-
- Bonaldi K, Gourion B, Fardoux J, Hannibal L, Cartieaux F et al. (2010). Large-scale transposon mutagenesis of photosynthetic Bradyrhizobium sp. strain ORS278 reveals new genetic loci putatively important for Nod-independent symbiosis with Aeschynomene indica. Mol Plant Microbe Interact 23: 760–770. - PubMed
-
- Chaintreuil C, Arrighi JF, Giraud E, Miché L, Moulin L, Dreyfus B et al. (2013). Evolution of symbiosis in the legume genus Aeschynomene. New Phytol 200: 1247–1259. - PubMed
-
- D'Haeze W, Holsters M. (2004). Surface polysaccharides enable bacteria to evade plant immunity. Trends Microbiol 12: 555–561. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

