The Origin and Evolution of Baeyer-Villiger Monooxygenases (BVMOs): An Ancestral Family of Flavin Monooxygenases
- PMID: 26161776
- PMCID: PMC4498894
- DOI: 10.1371/journal.pone.0132689
The Origin and Evolution of Baeyer-Villiger Monooxygenases (BVMOs): An Ancestral Family of Flavin Monooxygenases
Abstract
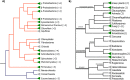
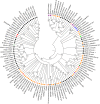
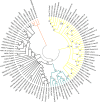
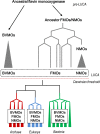
The Baeyer-Villiger Monooxygenases (BVMOs) are enzymes belonging to the "Class B" of flavin monooxygenases and are capable of performing exquisite selective oxidations. These enzymes have been studied from a biotechnological perspective, but their physiological substrates and functional roles are widely unknown. Here, we investigated the origin, taxonomic distribution and evolutionary history of the BVMO genes. By using in silico approaches, 98 BVMO encoding genes were detected in the three domains of life: Archaea, Bacteria and Eukarya. We found evidence for the presence of these genes in Metazoa (Hydra vulgaris, Oikopleura dioica and Adineta vaga) and Haptophyta (Emiliania huxleyi) for the first time. Furthermore, a search for other "Class B" monooxygenases (flavoprotein monooxygenases--FMOs--and N-hydroxylating monooxygenases--NMOs) was conducted. These sequences were also found in the three domains of life. Phylogenetic analyses of all "Class B" monooxygenases revealed that NMOs and BVMOs are monophyletic, whereas FMOs form a paraphyletic group. Based on these results, we propose that BVMO genes were already present in the last universal common ancestor (LUCA) and their current taxonomic distribution is the result of differential duplication and loss of paralogous genes.
Conflict of interest statement
Figures
References
-
- Gibson M, Nur-e-alam M, Lipata F, Oliveira MA, Rohr J. Characterization of kinetics and products of the Baeyer-Villiger oxygenase MtmOIV, the key enzyme of the biosynthetic pathway toward the natural product anticancer drug mithramycin from Streptomyces argillaceus. J Am Chem Soc. 2005;127(50):17594–5. Epub 2005/12/15. 10.1021/ja055750t . - DOI - PubMed
-
- Bosserman MA, Downey T, Noinaj N, Buchanan SK, Rohr J. Molecular insight into substrate recognition and catalysis of Baeyer-Villiger monooxygenase MtmOIV, the key frame-modifying enzyme in the biosynthesis of anticancer agent mithramycin. ACS Chem Biol. 2013;8(11):2466–77. Epub 2013/09/03. 10.1021/cb400399b - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

