Development of Recombinase Polymerase Amplification Assays for Detection of Orientia tsutsugamushi or Rickettsia typhi
- PMID: 26161793
- PMCID: PMC4498641
- DOI: 10.1371/journal.pntd.0003884
Development of Recombinase Polymerase Amplification Assays for Detection of Orientia tsutsugamushi or Rickettsia typhi
Abstract
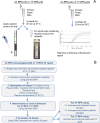
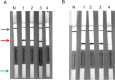
Sensitive, specific and rapid diagnostic tests for the detection of Orientia tsutsugamushi (O. tsutsugamushi) and Rickettsia typhi (R. typhi), the causative agents of scrub typhus and murine typhus, respectively, are necessary to accurately and promptly diagnose patients and ensure that they receive proper treatment. Recombinase polymerase amplification (RPA) assays using a lateral flow test (RPA-nfo) and real-time fluorescent detection (RPA-exo) were developed targeting the 47-kDa gene of O. tsutsugamushi or 17 kDa gene of R. typhi. The RPA assay was capable of detecting O. tsutsugamushi or R. typhi at levels comparable to that of the quantitative PCR method. Both the RPA-nfo and RPA-exo methods performed similarly with regards to sensitivity when detecting the 17 kDa gene of R. typhi. On the contrary, RPA-exo performed better than RPA-nfo in detecting the 47 kDa gene of O. tsutsugamushi. The clinical performance of the O. tsutsugamushi RPA assay was evaluated using either human patient samples or infected mouse samples. Eight out of ten PCR confirmed positives were determined positive by RPA, and all PCR confirmed negative samples were negative by RPA. Similar results were obtained for R. typhi spiked patient sera. The assays were able to differentiate O. tsutsugamushi and R. typhi from other phylogenetically related bacteria as well as mouse and human DNA. Furthermore, the RPA-nfo reaction was completed in 20 minutes at 37°C followed by a 10 minute incubation at room temperature for development of an immunochromatographic strip. The RPA-exo reaction was completed in 20 minutes at 39°C. The implementation of a cross contamination proof cassette to detect the RPA-nfo fluorescent amplicons provided an alternative to regular lateral flow detection strips, which are more prone to cross contamination. The RPA assays provide a highly time-efficient, sensitive and specific alternative to other methods for diagnosing scrub typhus or murine typhus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Development of a rapid and visual nucleotide detection method for a Chinese epidemic strain of Orientia tsutsugamushi based on recombinase polymerase amplification assay and lateral flow test.Int J Infect Dis. 2018 May;70:42-50. doi: 10.1016/j.ijid.2018.03.003. Epub 2018 Mar 13. Int J Infect Dis. 2018. PMID: 29548879
-
Establishment of Recombinase Polymerase Amplification assay for rapid and sensitive detection of Orientia tsutsugamushi in Southeast Asia.Acta Trop. 2020 Oct;210:105541. doi: 10.1016/j.actatropica.2020.105541. Epub 2020 May 31. Acta Trop. 2020. PMID: 32492397
-
Concurrent Infection with murine typhus and scrub typhus in southern Laos--the mixed and the unmixed.PLoS Negl Trop Dis. 2013 Aug 29;7(8):e2163. doi: 10.1371/journal.pntd.0002163. eCollection 2013. PLoS Negl Trop Dis. 2013. PMID: 24009783 Free PMC article. Review. No abstract available.
-
Serosurveillance of Orientia tsutsugamushi and Rickettsia typhi in Bangladesh.Am J Trop Med Hyg. 2014 Sep;91(3):580-583. doi: 10.4269/ajtmh.13-0570. Epub 2014 Aug 4. Am J Trop Med Hyg. 2014. PMID: 25092819 Free PMC article.
-
The typhus group.Clin Dermatol. 1996 May-Jun;14(3):271-8. doi: 10.1016/0738-081x(96)00011-9. Clin Dermatol. 1996. PMID: 8727129 Review. No abstract available.
Cited by
-
Assessment of a Sensitive qPCR Assay Targeting a Multiple-Copy Gene to Detect Orientia tsutsugamushi DNA.Trop Med Infect Dis. 2019 Jul 31;4(3):113. doi: 10.3390/tropicalmed4030113. Trop Med Infect Dis. 2019. PMID: 31370347 Free PMC article.
-
Development of a rapid and visual detection method for Rickettsia rickettsii combining recombinase polymerase assay with lateral flow test.PLoS One. 2018 Nov 26;13(11):e0207811. doi: 10.1371/journal.pone.0207811. eCollection 2018. PLoS One. 2018. PMID: 30475889 Free PMC article.
-
Rapid visual detection of anisakid nematodes using recombinase polymerase amplification and SYBR Green I.Front Microbiol. 2022 Dec 2;13:1026129. doi: 10.3389/fmicb.2022.1026129. eCollection 2022. Front Microbiol. 2022. PMID: 36532447 Free PMC article.
-
Recombinase polymerase amplification: Basics, applications and recent advances.Trends Analyt Chem. 2018 Jan;98:19-35. doi: 10.1016/j.trac.2017.10.015. Epub 2017 Oct 26. Trends Analyt Chem. 2018. PMID: 32287544 Free PMC article. Review.
-
Reducing Uncertainty for Acute Febrile Illness in Resource-Limited Settings: The Current Diagnostic Landscape.Am J Trop Med Hyg. 2017 Jun;96(6):1285-1295. doi: 10.4269/ajtmh.16-0667. Am J Trop Med Hyg. 2017. PMID: 28719277 Free PMC article. Review.
References
-
- Walker DH (2007) Rickettsiae and Rickettsial Infections: The Current State of Knowledge. Clin Infec Dis 45: S39–S44. - PubMed
-
- Bavaro MF, Kelly DJ, Dasch DA, Hale BR, Olson P (2005) History of U. S. Military Contributions to the study of Rickettsial Diseases. Military Medicine 170:49–60. - PubMed
-
- Kelly DJ, Richards AL, Temenak J, Strickman D, Dasch GA (2002) The past and present threat of rickettsial diseases to military medicine and international public health, Clinical Infect. Dis. 34: S145–S169. - PubMed
-
- Rolain JM, Jensenius M, Raoult D (2004) Rickettsial infections—a threat to travellers? Curr Opin Infect Dis 17:433–7. - PubMed
-
- Kovacova E, Kazar J (2000) Rickettsial diseases and their serological diagnosis. Clin Lab 46:239–45. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

