Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication
- PMID: 26162680
- PMCID: PMC4522766
- DOI: 10.1073/pnas.1503653112
Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication
Abstract
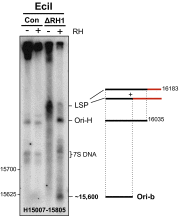
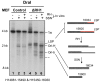
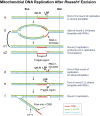
Encoding ribonuclease H1 (RNase H1) degrades RNA hybridized to DNA, and its function is essential for mitochondrial DNA maintenance in the developing mouse. Here we define the role of RNase H1 in mitochondrial DNA replication. Analysis of replicating mitochondrial DNA in embryonic fibroblasts lacking RNase H1 reveals retention of three primers in the major noncoding region (NCR) and one at the prominent lagging-strand initiation site termed Ori-L. Primer retention does not lead immediately to depletion, as the persistent RNA is fully incorporated in mitochondrial DNA. However, the retained primers present an obstacle to the mitochondrial DNA polymerase γ in subsequent rounds of replication and lead to the catastrophic generation of a double-strand break at the origin when the resulting gapped molecules are copied. Hence, the essential role of RNase H1 in mitochondrial DNA replication is the removal of primers at the origin of replication.
Keywords: DNA replication; RNase H; mitochondrial DNA; origin of replication; replication priming.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kasamatsu H, Vinograd J. Unidirectionality of replication in mouse mitochondrial DNA. Nat New Biol. 1973;241(108):103–105. - PubMed
-
- Crews S, Ojala D, Posakony J, Nishiguchi J, Attardi G. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature. 1979;277(5693):192–198. - PubMed
-
- Tapper DP, Clayton DA. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J Biol Chem. 1981;256(10):5109–5115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

