Most of the Dominant Members of Amphibian Skin Bacterial Communities Can Be Readily Cultured
- PMID: 26162880
- PMCID: PMC4561701
- DOI: 10.1128/AEM.01486-15
Most of the Dominant Members of Amphibian Skin Bacterial Communities Can Be Readily Cultured
Abstract
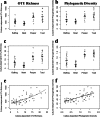
Currently, it is estimated that only 0.001% to 15% of bacteria in any given system can be cultured by use of commonly used techniques and media, yet culturing is critically important for investigations of bacterial function. Despite this situation, few studies have attempted to link culture-dependent and culture-independent data for a single system to better understand which members of the microbial community are readily cultured. In amphibians, some cutaneous bacterial symbionts can inhibit establishment and growth of the fungal pathogen Batrachochytrium dendrobatidis, and thus there is great interest in using these symbionts as probiotics for the conservation of amphibians threatened by B. dendrobatidis. The present study examined the portion of the culture-independent bacterial community (based on Illumina amplicon sequencing of the 16S rRNA gene) that was cultured with R2A low-nutrient agar and whether the cultured bacteria represented rare or dominant members of the community in the following four amphibian species: bullfrogs (Lithobates catesbeianus), eastern newts (Notophthalmus viridescens), spring peepers (Pseudacris crucifer), and American toads (Anaxyrus americanus). To determine which percentage of the community was cultured, we clustered Illumina sequences at 97% similarity, using the culture sequences as a reference database. For each amphibian species, we cultured, on average, 0.59% to 1.12% of each individual's bacterial community. However, the average percentage of bacteria that were culturable for each amphibian species was higher, with averages ranging from 2.81% to 7.47%. Furthermore, most of the dominant operational taxonomic units (OTUs), families, and phyla were represented in our cultures. These results open up new research avenues for understanding the functional roles of these dominant bacteria in host health.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures




References
-
- Ma L, Kim J, Hatzenpichler R, Karymov MA, Hubert N, Hanan IM, Chang EB, Ismagilov RF. 2014. Gene-targeted microfluidic cultivation validated by isolation of a gut bacterium listed in Human Microbiome Project's Most Wanted taxa. Proc Natl Acad Sci U S A 111:9768–9773. doi:10.1073/pnas.1404753111. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

