Rerouting Cellular Electron Flux To Increase the Rate of Biological Methane Production
- PMID: 26162885
- PMCID: PMC4561719
- DOI: 10.1128/AEM.01162-15
Rerouting Cellular Electron Flux To Increase the Rate of Biological Methane Production
Abstract
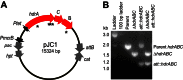
Methanogens are anaerobic archaea that grow by producing methane, a gas that is both an efficient renewable fuel and a potent greenhouse gas. We observed that overexpression of the cytoplasmic heterodisulfide reductase enzyme HdrABC increased the rate of methane production from methanol by 30% without affecting the growth rate relative to the parent strain. Hdr enzymes are essential in all known methane-producing archaea. They function as the terminal oxidases in the methanogen electron transport system by reducing the coenzyme M (2-mercaptoethane sulfonate) and coenzyme B (7-mercaptoheptanoylthreonine sulfonate) heterodisulfide, CoM-S-S-CoB, to regenerate the thiol-coenzymes for reuse. In Methanosarcina acetivorans, HdrABC expression caused an increased rate of methanogenesis and a decrease in metabolic efficiency on methylotrophic substrates. When acetate was the sole carbon and energy source, neither deletion nor overexpression of HdrABC had an effect on growth or methane production rates. These results suggest that in cells grown on methylated substrates, the cell compensates for energy losses due to expression of HdrABC with an increased rate of substrate turnover and that HdrABC lacks the appropriate electron donor in acetate-grown cells.
Copyright © 2015, Catlett et al.
Figures
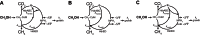
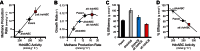

Similar articles
-
Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase.Mol Microbiol. 2010 Feb;75(4):843-53. doi: 10.1111/j.1365-2958.2009.06990.x. Epub 2009 Dec 4. Mol Microbiol. 2010. PMID: 19968794
-
A Ferredoxin- and F420H2-Dependent, Electron-Bifurcating, Heterodisulfide Reductase with Homologs in the Domains Bacteria and Archaea.mBio. 2017 Feb 7;8(1):e02285-16. doi: 10.1128/mBio.02285-16. mBio. 2017. PMID: 28174314 Free PMC article.
-
Supplementation of Sulfide or Acetate and 2-Mercaptoethane Sulfonate Restores Growth of the Methanosarcina acetivorans ΔhdrABC Deletion Mutant during Methylotrophic Methanogenesis.Microorganisms. 2023 Jan 28;11(2):327. doi: 10.3390/microorganisms11020327. Microorganisms. 2023. PMID: 36838292 Free PMC article.
-
Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens.Biochim Biophys Acta. 2014 Jul;1837(7):1130-47. doi: 10.1016/j.bbabio.2013.12.002. Epub 2013 Dec 12. Biochim Biophys Acta. 2014. PMID: 24333786 Review.
-
Redox-driven proton translocation in methanogenic Archaea.Cell Mol Life Sci. 2002 Sep;59(9):1513-33. doi: 10.1007/s00018-002-8526-3. Cell Mol Life Sci. 2002. PMID: 12440773 Free PMC article. Review.
Cited by
-
Genome-wide gene expression and RNA half-life measurements allow predictions of regulation and metabolic behavior in Methanosarcina acetivorans.BMC Genomics. 2016 Nov 16;17(1):924. doi: 10.1186/s12864-016-3219-8. BMC Genomics. 2016. PMID: 27852217 Free PMC article.
-
Reintegrating Biology Through the Nexus of Energy, Information, and Matter.Integr Comp Biol. 2022 Feb 5;61(6):2082-2094. doi: 10.1093/icb/icab174. Integr Comp Biol. 2022. PMID: 34374780 Free PMC article.
-
Metabolic shift at the class level sheds light on adaptation of methanogens to oxidative environments.ISME J. 2018 Feb;12(2):411-423. doi: 10.1038/ismej.2017.173. Epub 2017 Nov 14. ISME J. 2018. PMID: 29135970 Free PMC article.
-
Genome-Scale Metabolic Modeling of Archaea Lends Insight into Diversity of Metabolic Function.Archaea. 2017 Jan 4;2017:9763848. doi: 10.1155/2017/9763848. eCollection 2017. Archaea. 2017. PMID: 28133437 Free PMC article. Review.
-
Mer overexpression in Methanosarcina acetivorans affects growth and methanogenesis during substrate adaptation.Appl Environ Microbiol. 2025 May 21;91(5):e0067525. doi: 10.1128/aem.00675-25. Epub 2025 Apr 25. Appl Environ Microbiol. 2025. PMID: 40277366 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

