PLANT U-BOX PROTEIN10 Regulates MYC2 Stability in Arabidopsis
- PMID: 26163577
- PMCID: PMC4531359
- DOI: 10.1105/tpc.15.00385
PLANT U-BOX PROTEIN10 Regulates MYC2 Stability in Arabidopsis
Abstract
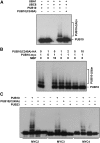
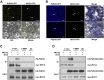
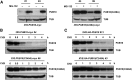
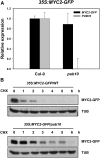
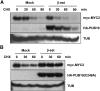
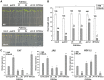
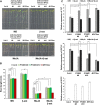
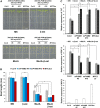
MYC2 is an important regulator for jasmonic acid (JA) signaling, but little is known about its posttranslational regulation. Here, we show that the MYC2 C-terminal region interacted with the PLANT U-BOX PROTEIN10 (PUB10) armadillo repeats in vitro. MYC2 was efficiently polyubiquitinated by PUB10 with UBC8 as an E2 enzyme and the conserved C249 in PUB10 was required for activity. The inactive PUB10(C249A) mutant protein retained its ability to heterodimerize with PUB10, thus blocking PUB10 E3 activity as a dominant-negative mutant. Both MYC2 and PUB10 were nucleus localized and coimmunoprecipitation experiments confirmed their interaction in vivo. Although unstable in the wild type, MYC2 stability was enhanced in pub10, suggesting destabilization by PUB10. Moreover, MYC2 half-life was shortened or prolonged by induced expression of PUB10 or the dominant-negative PUB10(C249A) mutant, respectively. Root growth of pub10 seedlings phenocopied 35S:MYC2 seedlings and was hypersensitive to methyl jasmonate, whereas 35S:PUB10 and jin1-9 (myc2) seedlings were hyposensitive. In addition, the root phenotype conferred by MYC2 overexpression in double transgenic plants was reversed or enhanced by induced expression of PUB10 or PUB10(C249A), respectively. Similar results were obtained with three other JA-regulated genes, TAT, JR2, and PDF1.2. Collectively, our results show that MYC2 is targeted by PUB10 for degradation during JA responses.
© 2015 American Society of Plant Biologists. All rights reserved.
Figures

Similar articles
-
The Deubiquitinating Enzymes UBP12 and UBP13 Positively Regulate MYC2 Levels in Jasmonate Responses.Plant Cell. 2017 Jun;29(6):1406-1424. doi: 10.1105/tpc.17.00216. Epub 2017 May 23. Plant Cell. 2017. PMID: 28536144 Free PMC article.
-
MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis.Plant Cell. 2007 Jul;19(7):2225-45. doi: 10.1105/tpc.106.048017. Epub 2007 Jul 6. Plant Cell. 2007. PMID: 17616737 Free PMC article.
-
A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis.Plant Cell. 2013 May;25(5):1641-56. doi: 10.1105/tpc.113.111112. Epub 2013 May 14. Plant Cell. 2013. PMID: 23673982 Free PMC article.
-
Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module.FEBS J. 2009 Sep;276(17):4682-92. doi: 10.1111/j.1742-4658.2009.07194.x. Epub 2009 Aug 3. FEBS J. 2009. PMID: 19663905 Review.
-
JAZ repressors set the rhythm in jasmonate signaling.Curr Opin Plant Biol. 2008 Oct;11(5):486-94. doi: 10.1016/j.pbi.2008.06.003. Epub 2008 Jul 22. Curr Opin Plant Biol. 2008. PMID: 18653378 Review.
Cited by
-
E3 Ubiquitin Ligases: Key Regulators of Hormone Signaling in Plants.Mol Cell Proteomics. 2018 Jun;17(6):1047-1054. doi: 10.1074/mcp.MR117.000476. Epub 2018 Mar 7. Mol Cell Proteomics. 2018. PMID: 29514858 Free PMC article. Review.
-
Wheat E3 ligase TaPRP19 is involved in drought stress tolerance in transgenic Arabidopsis.Physiol Mol Biol Plants. 2025 Feb;31(2):233-246. doi: 10.1007/s12298-025-01557-7. Epub 2025 Feb 10. Physiol Mol Biol Plants. 2025. PMID: 40070538
-
Jasmonic acid-responsive RRTF1 transcription factor controls DTX18 gene expression in hydroxycinnamic acid amide secretion.Plant Physiol. 2021 Mar 15;185(2):369-384. doi: 10.1093/plphys/kiaa043. Plant Physiol. 2021. PMID: 33721896 Free PMC article.
-
Measuring Protein Half-life in Arabidopsis thaliana.Bio Protoc. 2019 Aug 5;9(15):e3318. doi: 10.21769/BioProtoc.3318. eCollection 2019 Aug 5. Bio Protoc. 2019. PMID: 33654825 Free PMC article.
-
PUB11-Dependent Ubiquitination of the Phospholipid Flippase ALA10 Modifies ALA10 Localization and Affects the Pool of Linolenic Phosphatidylcholine.Front Plant Sci. 2020 Jul 15;11:1070. doi: 10.3389/fpls.2020.01070. eCollection 2020. Front Plant Sci. 2020. PMID: 32760418 Free PMC article.
References
-
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. - PubMed
-
- Andersen P., Kragelund B.B., Olsen A.N., Larsen F.H., Chua N.H., Poulsen F.M., Skriver K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279: 40053–40061. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

