mir-181a-1/b-1 Modulates Tolerance through Opposing Activities in Selection and Peripheral T Cell Function
- PMID: 26163591
- PMCID: PMC4763610
- DOI: 10.4049/jimmunol.1401587
mir-181a-1/b-1 Modulates Tolerance through Opposing Activities in Selection and Peripheral T Cell Function
Abstract
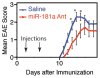
Understanding the consequences of tuning TCR signaling on selection, peripheral T cell function, and tolerance in the context of native TCR repertoires may provide insight into the physiological control of tolerance. In this study, we show that genetic ablation of a natural tuner of TCR signaling, mir-181a-1/b-1, in double-positive thymocytes dampened TCR and Erk signaling and increased the threshold of positive selection. Whereas mir-181a-1/b-1 deletion in mice resulted in an increase in the intrinsic reactivity of naive T cells to self-antigens, it did not cause spontaneous autoimmunity. Loss of mir-181a-1/b-1 dampened the induction of experimental autoimmune encephalomyelitis and reduced basal TCR signaling in peripheral T cells and their migration from lymph nodes to pathogenic sites. Taken together, these results demonstrate that tolerance can be modulated by microRNA gene products through the control of opposing activities in T cell selection and peripheral T cell function.
Copyright © 2015 by The American Association of Immunologists, Inc.
Figures
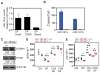



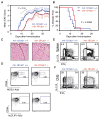
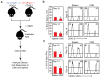
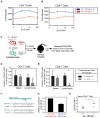
References
-
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. 1988;334:395–402. - PubMed
-
- Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. 2005;5:772–782. - PubMed
-
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. - PubMed
-
- Lo W-L, Allen PM. Self-Peptides in TCR Repertoire Selection and Peripheral T Cell Function. Current topics in microbiology and immunology 2013 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01HL107202/HL/NHLBI NIH HHS/United States
- P01 HL107202/HL/NHLBI NIH HHS/United States
- R01AI073724/AI/NIAID NIH HHS/United States
- DP1 CA174421/CA/NCI NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- R01 HL109102/HL/NHLBI NIH HHS/United States
- N01-HV-00242/HV/NHLBI NIH HHS/United States
- Canadian Institutes of Health Research/Canada
- DP1CA174421/CA/NCI NIH HHS/United States
- R01HL109102/HL/NHLBI NIH HHS/United States
- R01 AI073724/AI/NIAID NIH HHS/United States
- DP1 OD006435/OD/NIH HHS/United States
- 1U19AI100627/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

