Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum
- PMID: 26163699
- PMCID: PMC4588885
- DOI: 10.1093/jxb/erv340
Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum
Abstract
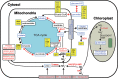
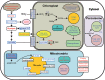
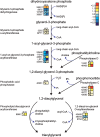
Algal growth is strongly affected by nitrogen (N) availability. Diatoms, an ecologically important group of unicellular algae, have evolved several acclimation mechanisms to cope with N deprivation. In this study, we integrated physiological data with transcriptional and metabolite data to reveal molecular and metabolic modifications in N-deprived conditions in the marine diatom Phaeodactylum tricornutum. Physiological and metabolite measurements indicated that the photosynthetic capacity and chlorophyll content of the cells decreased, while neutral lipids increased in N-deprived cultures. Global gene expression analysis showed that P. tricornutum responded to N deprivation through an increase in N transport, assimilation, and utilization of organic N resources. Following N deprivation, reduced biosynthesis and increased recycling of N compounds like amino acids, proteins, and nucleic acids was observed at the transcript level. The majority of the genes associated with photosynthesis and chlorophyll biosynthesis were also repressed. Carbon metabolism was restructured through downregulation of the Calvin cycle and chrysolaminarin biosynthesis, and co-ordinated upregulation of glycolysis, the tricarboxylic acid cycle, and pyruvate metabolism, leading to funnelling of carbon sources to lipid metabolism. Finally, reallocation of membrane lipids and induction of de novo triacylglycerol biosynthesis directed cells to accumulation of neutral lipids.
Keywords: Carbon metabolism; Phaeodactylum tricornutum; diatom; metabolomics; nitrogen deprivation; transcriptome; triacylglycerol..
© The Author 2015. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
-
- Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11, 36–43.
-
- Allen AE. 2005. Beyond sequence homology: redundant ammonium transporters in a marine diatom are not functionally equivalent. Journal of Phycology 41, 4–6.
-
- Allen AE, Dupont CL, Obornik M, et al. 2011. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207. - PubMed
-
- Allen AE, Ward BB, Song B. 2005. Characterization of diatom (Bacillariophyceae) nitrate reductase genes and their determination in marine phytoplankton communities. Journal of Phycology 41, 95–104.
-
- Allison G, Syrett PJ. 1987. The metabolism of guanine by the diatom Phaeodactylum tricornutum Bohlin. Journal of Phycology 23, 666–668.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

