Lipid insertion domain unfolding regulates protein orientational transition behavior in a lipid bilayer
- PMID: 26164502
- PMCID: PMC4554884
- DOI: 10.1016/j.bpc.2015.06.011
Lipid insertion domain unfolding regulates protein orientational transition behavior in a lipid bilayer
Abstract
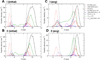
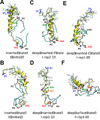
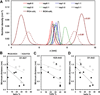
We have used coarse-grained (CG) and united atom (UA) molecular dynamics simulations to explore the mechanisms of protein orientational transition of a model peptide (Aβ42) in a phosphatidylcholine/cholesterol (PC/CHO) lipid bilayer. We started with an inserted state of Aβ42 containing a folded (I) or unfolded (II) K28-A42 lipid insertion domain (LID), which was stabilized by the K28-snorkeling and A42-anchoring to the PC polar groups in the lipid bilayer. After a UA-to-CG transformation and a 1000ns-CG simulation for enhancing the sampling of protein orientations, we discovered two transitions: I-to-"deep inserted" state with disrupted K28-snorkeling and II-to-"deep surface" state with disrupted A42-anchoring. The new states remained stable after a CG-to-UA transformation and a 200ns-UA simulation relaxation. Significant changes in the cholesterol-binding domain of Aβ42 and protein-induced membrane disruptions were evident after the transitions. We propose that the conformation of the LID regulates protein orientational transitions in the lipid membrane.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures








Similar articles
-
Cholesterol promotes the interaction of Alzheimer β-amyloid monomer with lipid bilayer.J Mol Biol. 2012 Aug 24;421(4-5):561-71. doi: 10.1016/j.jmb.2011.11.006. Epub 2011 Nov 15. J Mol Biol. 2012. PMID: 22108168
-
Molecular dynamics simulations reveal the protective role of cholesterol in β-amyloid protein-induced membrane disruptions in neuronal membrane mimics.J Phys Chem B. 2011 Aug 18;115(32):9795-812. doi: 10.1021/jp2012842. Epub 2011 Jul 26. J Phys Chem B. 2011. PMID: 21740063 Free PMC article.
-
Phase separation in a lipid/cholesterol system: comparison of coarse-grained and united-atom simulations.J Phys Chem B. 2013 Apr 11;117(14):3841-51. doi: 10.1021/jp312245y. Epub 2013 Mar 29. J Phys Chem B. 2013. PMID: 23470157
-
All-atom simulations and free-energy calculations of coiled-coil peptides with lipid bilayers: binding strength, structural transition, and effect on lipid dynamics.Sci Rep. 2016 Mar 1;6:22299. doi: 10.1038/srep22299. Sci Rep. 2016. PMID: 26926570 Free PMC article.
-
Molecular dynamics simulation studies of lipid bilayer systems.Acta Biochim Pol. 2000;47(3):601-11. Acta Biochim Pol. 2000. PMID: 11310963 Review.
Cited by
-
Exploring the Role of Anionic Lipid Nanodomains in the Membrane Disruption and Protein Folding of Human Islet Amyloid Polypeptide Oligomers on Lipid Membrane Surfaces Using Multiscale Molecular Dynamics Simulations.Molecules. 2023 May 19;28(10):4191. doi: 10.3390/molecules28104191. Molecules. 2023. PMID: 37241931 Free PMC article.
-
Characterization of Lipid-Protein Interactions and Lipid-Mediated Modulation of Membrane Protein Function through Molecular Simulation.Chem Rev. 2019 May 8;119(9):6086-6161. doi: 10.1021/acs.chemrev.8b00608. Epub 2019 Apr 12. Chem Rev. 2019. PMID: 30978005 Free PMC article. Review.
-
Data supporting beta-amyloid dimer structural transitions and protein-lipid interactions on asymmetric lipid bilayer surfaces using MD simulations on experimentally derived NMR protein structures.Data Brief. 2016 Mar 10;7:658-72. doi: 10.1016/j.dib.2016.03.015. eCollection 2016 Jun. Data Brief. 2016. PMID: 27054174 Free PMC article.
-
Maximally asymmetric transbilayer distribution of anionic lipids alters the structure and interaction with lipids of an amyloidogenic protein dimer bound to the membrane surface.Chem Phys Lipids. 2016 Mar;196:33-51. doi: 10.1016/j.chemphyslip.2016.01.002. Epub 2016 Jan 28. Chem Phys Lipids. 2016. PMID: 26827904 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

