Diet shapes the gut microbiome of pigs during nursing and weaning
- PMID: 26167280
- PMCID: PMC4499176
- DOI: 10.1186/s40168-015-0091-8
Diet shapes the gut microbiome of pigs during nursing and weaning
Abstract
Background: The newborn mammal is rapidly colonized by a complex microbial community, whose importance for host health is becoming increasingly clear. Understanding the forces that shape the early community, especially during the nursing period, is critical to gain insight into how this consortium of microbes is assembled. Pigs present an attractive model for nursing humans, given physiological and compositional similarity of pig and human milk and the utility of pigs in experimental studies. However, there is a paucity of data examining the gut microbiome in nursing pigs from birth through weaning using modern molecular methods and fewer experimental studies that examine the impact of diet on these microbial communities.
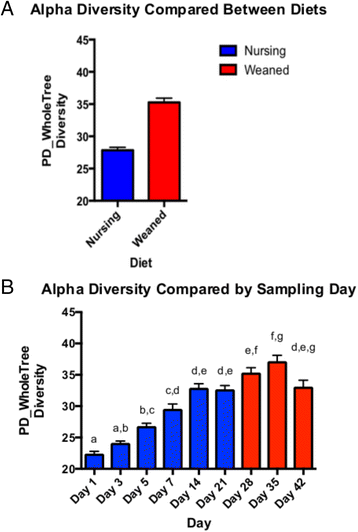
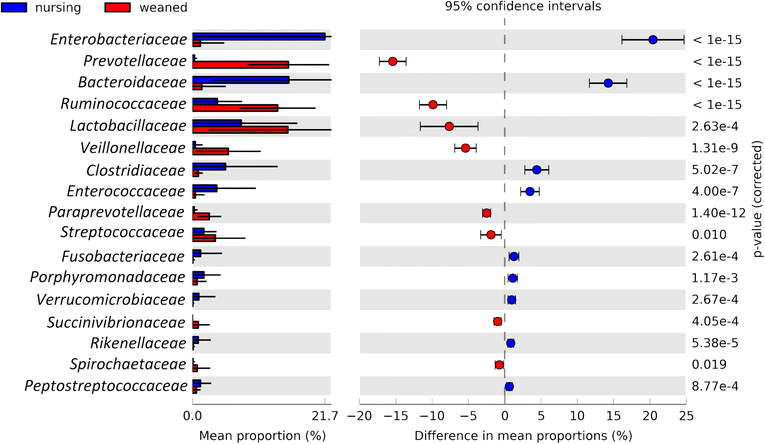
Results: We characterized the fecal microbiome of pigs from birth through 7 weeks of age, during which the animals were transitioned from an exclusive diet of sow milk to a starter diet composed of plant and animal-based components. Microbial communities were clearly distinguishable based on diet, being relatively stable absent dietary changes. Metagenomic sequencing was used to characterize a subset of animals before and after weaning, which identified glycan degradation pathways differing significantly between diets. Predicted enzymes active on milk-derived glycans that are otherwise indigestible to the host animal were enriched in the microbial metagenome of milk-fed animals. In contrast, the bacterial metagenome of weaned animals was enriched in functional pathways involved in plant glycan deconstruction and consumption.
Conclusions: The gut microbiome in young pigs is dramatically shaped by the composition of dietary glycans, reflected by the different functional capacities of the microbiome before and after weaning.
Figures




References
-
- Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014;15:811. doi: 10.1186/s13059-014-0552-6. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

