VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo
- PMID: 26167652
- PMCID: PMC4574298
- DOI: 10.7554/eLife.07366
VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo
Abstract
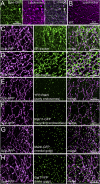
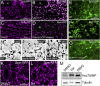
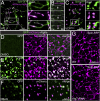
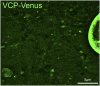
Lysosomes are classically viewed as vesicular structures to which cargos are delivered for degradation. Here, we identify a network of dynamic, tubular lysosomes that extends throughout Drosophila muscle, in vivo. Live imaging reveals that autophagosomes merge with tubular lysosomes and that lysosomal membranes undergo extension, retraction, fusion and fission. The dynamics and integrity of this tubular lysosomal network requires VCP, an AAA-ATPase that, when mutated, causes degenerative diseases of muscle, bone and neurons. We show that human VCP rescues the defects caused by loss of Drosophila VCP and overexpression of disease relevant VCP transgenes dismantles tubular lysosomes, linking tubular lysosome dysfunction to human VCP-related diseases. Finally, disruption of tubular lysosomes correlates with impaired autophagosome-lysosome fusion, increased cytoplasmic poly-ubiquitin aggregates, lipofuscin material, damaged mitochondria and impaired muscle function. We propose that VCP sustains sarcoplasmic proteostasis, in part, by controlling the integrity of a dynamic tubular lysosomal network.
Keywords: D. melanogaster; IBMPFD; amyotrophic lateral sclerosis; autophagy; cell biology; lysosome; neuroscience; skeletal muscle; spinster.
Conflict of interest statement
GWD: Reviewing editor,
The other authors declare that no competing interests exist.
Figures



References
-
- Abramzon Y, Johnson JO, Scholz SW, Taylor JP, Brunetti M, Calvo A, Mandrioli J, Benatar M, Mora G, Restagno G, Chiò A, Traynor BJ. Valosin-containing protein (VCP) mutations in sporadic amyotrophic lateral sclerosis. Neurobiology of Aging. 2012;33:2231. doi: 10.1016/j.neurobiolaging.2012.04.005. - DOI - PMC - PubMed
-
- Chang YC, Hung WT, Chang YC, Chang HC, Wu CL, Chiang AS, Jackson GR, Sang TK. Pathogenic VCP/TER94 alleles are dominant actives and contribute to neurodegeneration by altering cellular ATP level in a Drosophila IBMPFD model. PLOS Genetics. 2011;7:e1001288. doi: 10.1371/journal.pgen.1001288. - DOI - PMC - PubMed
-
- Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proceedings of the National Academy of Sciences of USA. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. - DOI - PMC - PubMed
-
- Clemen CS, Tangavelou K, Strucksberg KH, Just S, Gaertner L, Regus-Leidig H, Stumpf M, Reimann J, Coras R, Morgan RO, Fernandez MP, Hofmann A, Müller S, Schoser B, Hanisch FG, Rottbauer W, Blümcke I, von Hörsten S, Eichinger L, Schröder R. Strumpellin is a novel valosin-containing protein binding partner linking hereditary spastic paraplegia to protein aggregation diseases. Brain. 2010;133:2920–2941. doi: 10.1093/brain/awq222. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

