Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk
- PMID: 26168013
- PMCID: PMC4930791
- DOI: 10.1038/ng.3353
Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk
Abstract
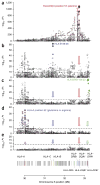
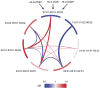
Variation in the human leukocyte antigen (HLA) genes accounts for one-half of the genetic risk in type 1 diabetes (T1D). Amino acid changes in the HLA-DR and HLA-DQ molecules mediate most of the risk, but extensive linkage disequilibrium complicates the localization of independent effects. Using 18,832 case-control samples, we localized the signal to 3 amino acid positions in HLA-DQ and HLA-DR. HLA-DQβ1 position 57 (previously known; P = 1 × 10(-1,355)) by itself explained 15.2% of the total phenotypic variance. Independent effects at HLA-DRβ1 positions 13 (P = 1 × 10(-721)) and 71 (P = 1 × 10(-95)) increased the proportion of variance explained to 26.9%. The three positions together explained 90% of the phenotypic variance in the HLA-DRB1-HLA-DQA1-HLA-DQB1 locus. Additionally, we observed significant interactions for 11 of 21 pairs of common HLA-DRB1-HLA-DQA1-HLA-DQB1 haplotypes (P = 1.6 × 10(-64)). HLA-DRβ1 positions 13 and 71 implicate the P4 pocket in the antigen-binding groove, thus pointing to another critical protein structure for T1D risk, in addition to the HLA-DQ P9 pocket.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

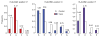
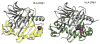

References
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK062418/DK/NIDDK NIH HHS/United States
- 5R01AR062886-02/AR/NIAMS NIH HHS/United States
- UH2 AR067677/AR/NIAMS NIH HHS/United States
- R01 AR065183/AR/NIAMS NIH HHS/United States
- 100140/WT_/Wellcome Trust/United Kingdom
- 1UH2AR067677-01/AR/NIAMS NIH HHS/United States
- 1R01AR063759/AR/NIAMS NIH HHS/United States
- 5U01GM092691-05/GM/NIGMS NIH HHS/United States
- RG/08/014/24067/BHF_/British Heart Foundation/United Kingdom
- DH_/Department of Health/United Kingdom
- R01AR065183/AR/NIAMS NIH HHS/United States
- MR/L003120/1/MRC_/Medical Research Council/United Kingdom
- U01 GM092691/GM/NIGMS NIH HHS/United States
- R01 AR063759/AR/NIAMS NIH HHS/United States
- R01 AR062886/AR/NIAMS NIH HHS/United States
- 091157/WT_/Wellcome Trust/United Kingdom
- U01DK062418/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

