Intradomain Confinement of Disulfides in the Folding of Two Consecutive Modules of the LDL Receptor
- PMID: 26168158
- PMCID: PMC4500599
- DOI: 10.1371/journal.pone.0132141
Intradomain Confinement of Disulfides in the Folding of Two Consecutive Modules of the LDL Receptor
Abstract
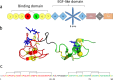
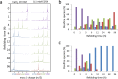
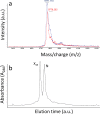
The LDL receptor internalizes circulating LDL and VLDL particles for degradation. Its extracellular binding domain contains ten (seven LA and three EGF) cysteine-rich modules, each bearing three disulfide bonds. Despite the enormous number of disulfide combinations possible, LDLR oxidative folding leads to a single native species with 30 unique intradomain disulfides. Previous folding studies of the LDLR have shown that non native disulfides are initially formed that lead to compact species. Accordingly, the folding of the LDLR has been described as a "coordinated nonvectorial" reaction, and it has been proposed that early compaction funnels the reaction toward the native structure. Here we analyze the oxidative folding of LA4 and LA5, the modules critical for ApoE binding, isolated and in the LA45 tandem. Compared to LA5, LA4 folding is slow and inefficient, resembling that of LA5 disease-linked mutants. Without Ca++, it leads to a mixture of many two-disulfide scrambled species and, with Ca++, to the native form plus two three-disulfide intermediates. The folding of the LA45 tandem seems to recapitulate that of the individual repeats. Importantly, although the folding of the LA45 tandem takes place through formation of scrambled isomers, no interdomain disulfides are detected, i.e. the two adjacent modules fold independently without the assistance of interdomain covalent interactions. Reduction of incredibly large disulfide combinatorial spaces, such as that in the LDLR, by intradomain confinement of disulfide bond formation might be also essential for the efficient folding of other homologous disulfide-rich receptors.
Conflict of interest statement
Figures





References
-
- Flory PJ. Theory of elastic mechanisms in fibrous proteins. J Am Chem Soc 1956;78:5222–35.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

