The KLP-7 Residue S546 Is a Putative Aurora Kinase Site Required for Microtubule Regulation at the Centrosome in C. elegans
- PMID: 26168236
- PMCID: PMC4500558
- DOI: 10.1371/journal.pone.0132593
The KLP-7 Residue S546 Is a Putative Aurora Kinase Site Required for Microtubule Regulation at the Centrosome in C. elegans
Abstract
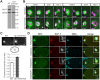
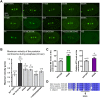
Regulation of microtubule dynamics is essential for many cellular processes, including proper assembly and function of the mitotic spindle. The kinesin-13 microtubule-depolymerizing enzymes provide one mechanism to regulate microtubule behaviour temporally and spatially. Vertebrate MCAK locates to chromatin, kinetochores, spindle poles, microtubule tips, and the cytoplasm, implying that the regulation of kinesin-13 activity and subcellular targeting is complex. Phosphorylation of kinesin-13 by Aurora kinase inhibits microtubule depolymerization activity and some Aurora phosphorylation sites on kinesin-13 are required for subcellular localization. Herein, we determine that a C. elegans deletion mutant klp-7(tm2143) causes meiotic and mitotic defects that are consistent with an increase in the amount of microtubules in the cytoplasmic and spindle regions of meiotic embryos, and an increase in microtubules emanating from centrosomes. We show that KLP-7 is phosphorylated by Aurora A and Aurora B kinases in vitro, and that the phosphorylation by Aurora A is stimulated by TPXL-1. Using a structure-function approach, we establish that one putative Aurora kinase site, S546, within the C-terminal part of the core domain is required for the function, but not subcellular localization, of KLP-7 in vivo. Furthermore, FRAP analysis reveals microtubule-dependent differences in the turnover of KLP-7(S546A) and KLP-7(S546E) mutant proteins at the centrosome, suggesting a possible mechanism for the regulation of KLP-7 by Aurora kinase.
Conflict of interest statement
Figures





References
-
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, et al. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(7):2741–6. Epub 2011/02/02. 10.1073/pnas.1016498108 - DOI - PMC - PubMed
-
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96(1):69–78. Epub 1999/02/16. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

