IncA/C Plasmid Carrying bla(NDM-1), bla(CMY-16), and fosA3 in a Salmonella enterica Serovar Corvallis Strain Isolated from a Migratory Wild Bird in Germany
- PMID: 26169417
- PMCID: PMC4576047
- DOI: 10.1128/AAC.00944-15
IncA/C Plasmid Carrying bla(NDM-1), bla(CMY-16), and fosA3 in a Salmonella enterica Serovar Corvallis Strain Isolated from a Migratory Wild Bird in Germany
Abstract
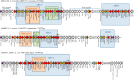
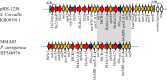
A Salmonella enterica serovar Corvallis strain was isolated from a wild bird in Germany. This strain carried the IncA/C2 pRH-1238 plasmid. Complete sequencing of the plasmid was performed, identifying the blaNDM-1, blaCMY-16, fosA3, sul1, sul2, strA, strB, aac(6')-Ib, aadA5, aphA6, tetA(A), mphA, floR, dfrA7, and merA genes, which confer clinically relevant resistance to most of the antimicrobial classes, including β-lactams with carbapenems, fosfomycin, aminoglycosides, co-trimoxazole, tetracyclines, and macrolides. The strain likely originated from the Asiatic region and was transferred to Germany through the Milvus migrans migratory route.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Genomic characterization of a large plasmid containing a bla NDM-1 gene carried on Salmonella enterica serovar Indiana C629 isolate from China.BMC Infect Dis. 2017 Jul 7;17(1):479. doi: 10.1186/s12879-017-2515-5. BMC Infect Dis. 2017. PMID: 28687066 Free PMC article.
-
Analysis of antimicrobial resistance genes detected in multidrug-resistant Salmonella enterica serovar Typhimurium isolated from food animals.Microb Drug Resist. 2011 Sep;17(3):407-18. doi: 10.1089/mdr.2010.0189. Epub 2011 Apr 3. Microb Drug Resist. 2011. PMID: 21457076
-
NDM-1 carbapenemase-producing Salmonella enterica subsp. enterica serovar Corvallis isolated from a wild bird in Germany.J Antimicrob Chemother. 2013 Dec;68(12):2954-6. doi: 10.1093/jac/dkt260. Epub 2013 Jun 30. J Antimicrob Chemother. 2013. PMID: 23818284 No abstract available.
-
Characterization of bla(CMY)-encoding plasmids among Salmonella isolated in the United States in 2007.Foodborne Pathog Dis. 2011 Dec;8(12):1289-94. doi: 10.1089/fpd.2011.0944. Epub 2011 Sep 1. Foodborne Pathog Dis. 2011. PMID: 21883005
-
[Phenotypic and molecular characteristics of Salmonella enterica serotype 1,4, [5] ,12:i:--a review].Wei Sheng Wu Xue Bao. 2014 Nov 4;54(11):1248-55. Wei Sheng Wu Xue Bao. 2014. PMID: 25752131 Review. Chinese.
Cited by
-
Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli.J Antimicrob Chemother. 2016 Sep;71(9):2460-5. doi: 10.1093/jac/dkw177. Epub 2016 Jun 3. J Antimicrob Chemother. 2016. PMID: 27261267 Free PMC article.
-
Characterization of a fosA3 Carrying IncC-IncN Plasmid From a Multidrug-Resistant ST17 Salmonella Indiana Isolate.Front Microbiol. 2020 Jul 21;11:1582. doi: 10.3389/fmicb.2020.01582. eCollection 2020. Front Microbiol. 2020. PMID: 32793137 Free PMC article.
-
Characterization of NDM-Encoding Plasmids From Enterobacteriaceae Recovered From Czech Hospitals.Front Microbiol. 2018 Jul 10;9:1549. doi: 10.3389/fmicb.2018.01549. eCollection 2018. Front Microbiol. 2018. PMID: 30042758 Free PMC article.
-
Emergence of Colistin Resistance Gene mcr-1 in Cronobacter sakazakii Producing NDM-9 and in Escherichia coli from the Same Animal.Antimicrob Agents Chemother. 2017 Jan 24;61(2):e01444-16. doi: 10.1128/AAC.01444-16. Print 2017 Feb. Antimicrob Agents Chemother. 2017. PMID: 27855074 Free PMC article.
-
Characterization of blaKPC-2 and blaNDM-1 Plasmids of a K. pneumoniae ST11 Outbreak Clone.Antibiotics (Basel). 2023 May 18;12(5):926. doi: 10.3390/antibiotics12050926. Antibiotics (Basel). 2023. PMID: 37237829 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

