The role of climate and out-of-Africa migration in the frequencies of risk alleles for 21 human diseases
- PMID: 26170196
- PMCID: PMC4501093
- DOI: 10.1186/s12863-015-0239-3
The role of climate and out-of-Africa migration in the frequencies of risk alleles for 21 human diseases
Abstract
Background: Demography and environmental adaptation can affect the global distribution of genetic variants and possibly the distribution of disease. Population heterozygosity of single nucleotide polymorphisms has been shown to decrease strongly with distance from Africa and this has been attributed to the effect of serial founding events during the migration of humans out of Africa. Additionally, population allele frequencies have been shown to change due to environmental adaptation. Here, we investigate the relationship of Out-of-Africa migration and climatic variables to the distribution of risk alleles for 21 diseases.
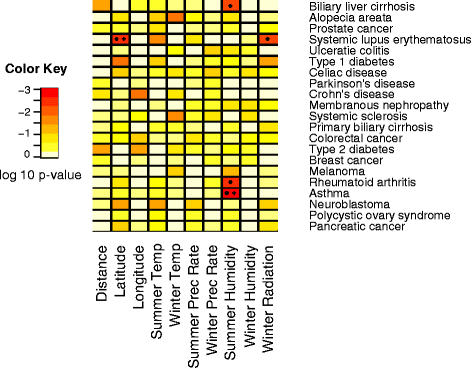
Results: For each disease, we computed the regression of average heterozygosity and average allele frequency of the risk alleles with distance from Africa and 9 environmental variables. We compared these regressions to a null distribution created by regressing statistics for SNPs not associated with disease on distance from Africa and these environmental variables. Additionally, we used Bayenv 2.0 to assess the signal of environmental adaptation associated with individual risk SNPs. For those SNPs in HGDP and HapMap that are risk alleles for type 2 diabetes, we cannot reject that their distribution is as expected from Out-of-Africa migration. However, the allelic statistics for many other diseases correlate more closely with environmental variables than would be expected from the serial founder effect and show signals of environmental adaptation. We report strong environmental interactions with several autoimmune diseases, and note a particularly strong interaction between asthma and summer humidity. Additionally, we identified several risk genes with strong environmental associations.
Conclusions: For most diseases, migration does not explain the distribution of risk alleles and the worldwide pattern of allele frequencies for some diseases may be better explained by environmental associations, which suggests that some selection has acted on these diseases.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

