Mycobacterium smegmatis HelY Is an RNA-Activated ATPase/dATPase and 3'-to-5' Helicase That Unwinds 3'-Tailed RNA Duplexes and RNA:DNA Hybrids
- PMID: 26170411
- PMCID: PMC4560288
- DOI: 10.1128/JB.00418-15
Mycobacterium smegmatis HelY Is an RNA-Activated ATPase/dATPase and 3'-to-5' Helicase That Unwinds 3'-Tailed RNA Duplexes and RNA:DNA Hybrids
Abstract
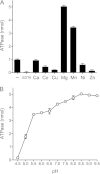
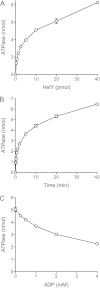
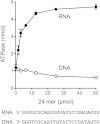
Mycobacteria have a large and distinctive ensemble of DNA helicases that function in DNA replication, repair, and recombination. Little is known about the roster of RNA helicases in mycobacteria or their roles in RNA transactions. The 912-amino-acid Mycobacterium smegmatis HelY (MSMEG_3885) protein is a bacterial homolog of the Mtr4 and Ski2 helicases that regulate RNA 3' processing and turnover by the eukaryal exosome. Here we characterize HelY as an RNA-stimulated ATPase/dATPase and an ATP/dATP-dependent 3'-to-5' helicase. HelY requires a 3' single-strand RNA tail (a loading RNA strand) to displace the complementary strand of a tailed RNA:RNA or RNA:DNA duplex. The findings that HelY ATPase is unresponsive to a DNA polynucleotide cofactor and that HelY is unable to unwind a 3'-tailed duplex in which the loading strand is DNA distinguish HelY from other mycobacterial nucleoside triphosphatases/helicases characterized previously. The biochemical properties of HelY, which resemble those of Mtr4/Ski2, hint at a role for HelY in mycobacterial RNA catabolism.
Importance: RNA helicases play crucial roles in transcription, RNA processing, and translation by virtue of their ability to alter RNA secondary structure or remodel RNA-protein interactions. In eukarya, the RNA helicases Mtr4 and Ski2 regulate RNA 3' resection by the exosome. Mycobacterium smegmatis HelY, a bacterial homolog of Mtr4/Ski2, is characterized here as a unidirectional helicase, powered by RNA-dependent ATP/dATP hydrolysis, that tracks 3' to 5' along a loading RNA strand to displace the complementary strand of a tailed RNA:RNA or RNA:DNA duplex. The biochemical properties of HelY suggest a role in bacterial RNA transactions. HelY homologs are present in pathogenic mycobacteria (e.g., M. tuberculosis and M. leprae) and are widely prevalent in Actinobacteria and Cyanobacteria but occur sporadically elsewhere in the bacterial domain.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





Similar articles
-
Mycobacterium smegmatis Lhr Is a DNA-dependent ATPase and a 3'-to-5' DNA translocase and helicase that prefers to unwind 3'-tailed RNA:DNA hybrids.J Biol Chem. 2013 May 17;288(20):14125-14134. doi: 10.1074/jbc.M113.466854. Epub 2013 Apr 2. J Biol Chem. 2013. PMID: 23549043 Free PMC article.
-
The mycobacterial PhoH2 proteins are type II toxin antitoxins coupled to RNA helicase domains.Tuberculosis (Edinb). 2015 Jul;95(4):385-94. doi: 10.1016/j.tube.2015.03.013. Epub 2015 Apr 16. Tuberculosis (Edinb). 2015. PMID: 25999286
-
Biochemical Characterization of Mycobacterium smegmatis RnhC (MSMEG_4305), a Bifunctional Enzyme Composed of Autonomous N-Terminal Type I RNase H and C-Terminal Acid Phosphatase Domains.J Bacteriol. 2015 Aug 1;197(15):2489-98. doi: 10.1128/JB.00268-15. Epub 2015 May 18. J Bacteriol. 2015. PMID: 25986906 Free PMC article.
-
Ski2-like RNA helicase structures: common themes and complex assemblies.RNA Biol. 2013 Jan;10(1):33-43. doi: 10.4161/rna.22101. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995828 Free PMC article. Review.
-
Structural aspects of RNA helicases in eukaryotic mRNA decay.Biosci Rep. 2009 Jul 13;29(5):339-49. doi: 10.1042/BSR20090034. Biosci Rep. 2009. PMID: 19589129 Review.
Cited by
-
Genome-Wide Transcriptional Response of Mycobacterium smegmatis MC2155 to G-Quadruplex Ligands BRACO-19 and TMPyP4.Front Microbiol. 2022 Mar 4;13:817024. doi: 10.3389/fmicb.2022.817024. eCollection 2022. Front Microbiol. 2022. PMID: 35308348 Free PMC article.
-
A research program-linked, course-based undergraduate research experience that allows undergraduates to participate in current research on mycobacterial gene regulation.Front Microbiol. 2023 Jan 6;13:1025250. doi: 10.3389/fmicb.2022.1025250. eCollection 2022. Front Microbiol. 2023. PMID: 36687599 Free PMC article.
-
RNases and Helicases in Gram-Positive Bacteria.Microbiol Spectr. 2018 Apr;6(2):10.1128/microbiolspec.rwr-0003-2017. doi: 10.1128/microbiolspec.RWR-0003-2017. Microbiol Spectr. 2018. PMID: 29651979 Free PMC article.
-
Small RNA Mcr11 requires the transcription factor AbmR for stable expression and regulates genes involved in the central metabolism of Mycobacterium tuberculosis.Mol Microbiol. 2020 Feb;113(2):504-520. doi: 10.1111/mmi.14436. Epub 2019 Dec 16. Mol Microbiol. 2020. PMID: 31782837 Free PMC article.
-
Immunoprecipitation of RNA-DNA hybrid interacting proteins in Trypanosoma brucei reveals conserved and novel activities, including in the control of surface antigen expression needed for immune evasion by antigenic variation.Nucleic Acids Res. 2023 Nov 10;51(20):11123-11141. doi: 10.1093/nar/gkad836. Nucleic Acids Res. 2023. PMID: 37843098 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

